Breast Histopathology Image Segmentation Part 1

- Part 1: Data Inspection and Pre-processing
- Part 2: Weights, Data Augmentations and Generators
- Part 3: Model creation based on a pre-trained and a custom model
- Part 4: Train our model to fit the dataset
- Part 5: Evaluate the performance of your trained model
- Part 6: Running Predictions
Based on Breast Histopathology Images by Paul Mooney.
Invasive Ductal Carcinoma (IDC) is the most common subtype of all breast cancers. To assign an aggressiveness grade to a whole mount sample, pathologists typically focus on the regions which contain the IDC. As a result, one of the common pre-processing steps for automatic aggressiveness grading is to delineate the exact regions of IDC inside of a whole mount slide.Can recurring breast cancer be spotted with AI tech? - BBC News
- Citation: Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases
- Dataset: 198,738 IDC(negative) image patches; 78,786 IDC(positive) image patches
Data Preprocessing
After downloading the source photographs unzip them into a folder ./dataset/raw. Now we need to process this data and process it for the model training.
Create the Training, Test and Validation Set
The configuration file defines the location where data will be stored as well as setting the parameter for the model training:
utils/config.py
import os
# Project location
PROJECT_PATH = "./"
# Input dataset directory
RAW_INPUT_PATH = "./dataset/raw"
# Result dataset directory
SPLIT_INPUT_PATH = "./dataset/split"
# Output directory
OUTPUT_PATH = "./output"
# Training testing, validation
TRAIN_PATH = os.path.sep.join([SPLIT_INPUT_PATH, "training"])
VAL_PATH = os.path.sep.join([SPLIT_INPUT_PATH, "validation"])
TEST_PATH = os.path.sep.join([SPLIT_INPUT_PATH, "testing"])
# Data splitting
TRAIN_SPLIT = 0.8
VAL_SPLIT = 0.1
# Parameters
CLASSES = ["benign","malignant"]
BATCH_SIZE = 32
INIT_LR = 1e-4
EPOCHS = 20
# Path to serialized model after training
MODEL_PATH = os.path.sep.join([OUTPUT_PATH, "CarcinomaPrediction.model"])
# Path to training history plots
MODEL_PATH = os.path.sep.join([OUTPUT_PATH, "TrainingHistory.png"])
First we need to split the raw dataset into training and validation set, according to the split ratio set above:
utils/create_dataset.py
import config
import getPaths
import shutil
import random
import os
# Get content of the original input directory and shuffle images
allImagePaths = list(getPaths.listImages(config.RAW_INPUT_PATH))
random.seed(42)
random.shuffle(allImagePaths)
# Only take 10% of the images to speed up the process
print(len(allImagePaths))
imagePaths = allImagePaths[0:20000]
print(len(allImagePaths))
# Split into training and testing data
i = int(len(allImagePaths) * config.TRAIN_SPLIT)
trainPaths = allImagePaths[:i]
testPaths = allImagePaths[i:]
# Separate validation split from training data
i = int(len(trainPaths) * config.VAL_SPLIT)
valPaths = trainPaths[:i]
trainPaths = trainPaths[i:]
# Defining the datasets which will be build in the result folder
datasets = [
("training", config.TRAIN_PATH, trainPaths),
("validation", config.VAL_PATH, valPaths),
("testing", config.TEST_PATH, testPaths)
]
# Copy images from the initial into the result path
# while splitting them into train, validation and test data
for (dSType, basePath, allImagePaths) in datasets:
print("Making '{}' split".format(dSType))
if not os.path.exists(basePath):
print("'Creating {}' directory".format(basePath))
os.makedirs(basePath)
# Looping over the image paths
for inputPath in allImagePaths:
# Extracting the filename of the input image
# Extracting class label ("0" for "Benign" and "1" for "Malignant")
filename = inputPath.split(os.path.sep)[-1]
label = filename[-5:-4]
# Making the path to form the label directory
labelPath = os.path.sep.join([basePath, label])
if not os.path.exists(labelPath):
print("'creating {}' directory".format(labelPath))
os.makedirs(labelPath)
# Creating the path to the destination image and then copying it
finalPath = os.path.sep.join([labelPath, filename])
shutil.copy2(inputPath, finalPath)
This file imports a helper function getPaths that helps us decide what files inside the dataset we actually want to use based on their file extension:
utils/getPaths.py
import os
# Defining image types you want to allow
imageTypes = (".jpg", ".jpeg", ".png", ".tif", ".tiff", ".bmp")
def listImages(basePath, contains=None):
return listFiles(basePath, validExts=imageTypes, contains=contains)
def listFiles(basePath, validExts=None, contains=None):
for (baseDir, dirNames, filenames) in os.walk(basePath):
for filename in filenames:
# Get all files in filename / ignore empty directories
if contains is not None and filename.find(contains) == -1:
continue
# Extracting the file extension
fileExt = filename[filename.rfind("."):].lower()
# Only process files that are of imageTypes
if validExts is None or fileExt.endswith(validExts):
# Construct the path to the image and yield it
imagePath = os.path.join(baseDir, filename)
yield imagePath
Running the script:
python ./utils/create_dataset.py
Making 'training' split
'Creating ./dataset/split/training' directory
'creating ./dataset/split/training/0' directory
'creating ./dataset/split/training/1' directory
Making 'validation' split
'Creating ./dataset/split/validation' directory
'creating ./dataset/split/validation/0' directory
'creating ./dataset/split/validation/1' directory
Making 'testing' split
'Creating ./dataset/split/testing' directory
'creating ./dataset/split/testing/0' directory
'creating ./dataset/split/testing/1' directory
will now go through the raw images in ./dataset/raw and split them into the ./dataset/split folders:
.
├── raw
├── split
│ ├── testing
| │ ├── 0
| │ └── 1
│ ├── training
| │ ├── 0
| │ └── 1
│ └── validation
| │ ├── 0
| │ └── 1
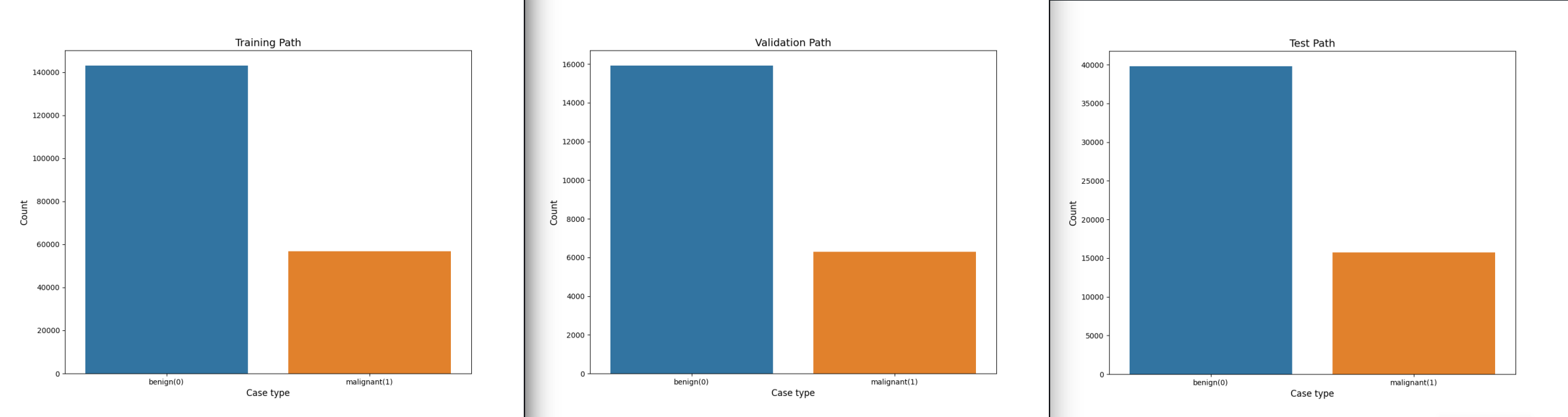
We will get a 80% training, 10% testing and 10% validation split. And based on the file names the images will be separated into benign 0 and malignant 1.
Custom Model Building and Evaluation
Requirements
Install the dependencies using pip or pipenv:
pipenv install tensorflow-gpu scikit-learn matplotlib numpy pandas seaborn opencv-python
or
pip install -r requirements.txt --upgrade
Exploring the Dataset
Distribution
utils/plotDistribution.py
# Plotting the count of images within each segment in a directories
def plotData(dirPath):
# Get the path to the benign and malignant sub-directories
benign_cases_dir = dirPath+ '/0/'
malignant_cases_dir = dirPath + '/1/'
# Get the list of all the images from those paths
benign_cases = glob.glob(benign_cases_dir + '*.png')
malignant_cases = glob.glob(malignant_cases_dir + '*.png')
# An empty list
data1 = []
# Add all benign cases to list with label `0`
for img in benign_cases:
data1.append((img,0))
# Add all benign cases to list with label `1`
for img in malignant_cases:
data1.append((img, 1))
# data => pandas dataframe
data1 = pd.DataFrame(data1, columns=['image', 'label'],index=None)
# Shuffle the data
data1 = data1.sample(frac=1.).reset_index(drop=True)
# Get the counts for each segment
cases_count = data1['label'].value_counts()
print(cases_count)
# Plot the results
plt.figure(figsize=(10,8))
sns.barplot(x=cases_count.index, y= cases_count.values)
plt.title('Number of cases', fontsize=14)
plt.xlabel('Case type', fontsize=12)
plt.ylabel('Count', fontsize=12)
plt.xticks(range(len(cases_count.index)), ['benign(0)', 'malignant(1)'])
plt.show()
try:
function = sys.argv[1]
globals()[function]()
except IndexError:
raise Exception("Please provide function name")
except KeyError:
raise Exception("Function {} hasn't been found".format(function))
By importing the configuration file from utils import config we can now print out the distribution of benign and malignant cases in all three folders by running the following commands after adding a little helper function:
def exploreData():
plotData(config.TRAIN_PATH,"Training Path")
# plotData(config.VAL_PATH,"Validation Path")
# plotData(config.TEST_PATH,"Test Path")
The function can be executed directly:
pipenv run python ./utils/plotDistribution.py exploreData
We can see that we are working with a skewed dataset that is heavy on benign cases. But the splitting of images into the three paths was successful - we did not change the distribution:
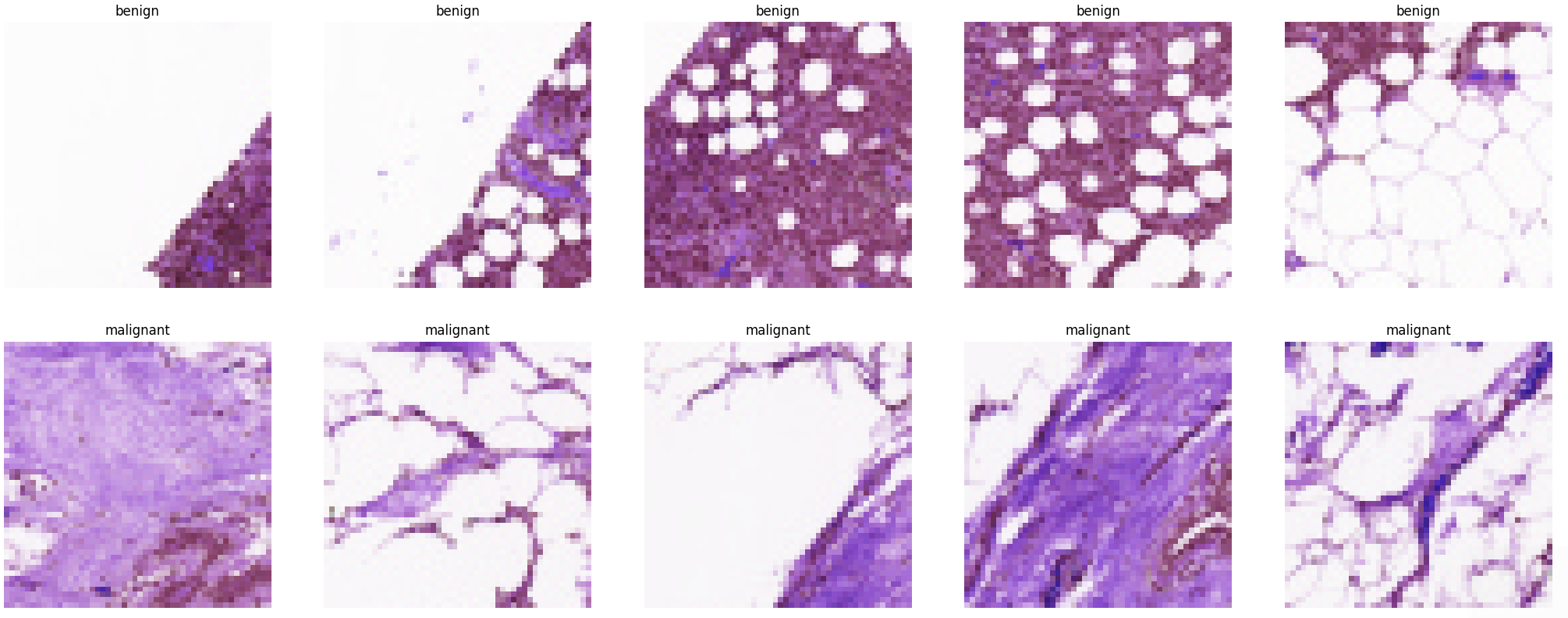
Sample Images
Now we can inspect our data by printing a sample of each class:
# Get the path to the benign and malignant sub-directories
benign_cases_dir = config.TRAIN_PATH + '/0/'
malignant_cases_dir = config.TRAIN_PATH + '/1/'
# Get the list of all the images from those paths
benign_cases = glob.glob(benign_cases_dir + '*.png')
malignant_cases = glob.glob(malignant_cases_dir + '*.png')
# An empty list
train_data1 = []
## Add all benign cases to list with label `0`
for img in benign_cases:
train_data1.append((img,0))
# Go through all the malignant cases. The label for these cases will be 1
for img in malignant_cases:
train_data1.append((img, 1))
# Add all benign cases to list with label `1`
train_data1 = pd.DataFrame(train_data1, columns=['image', 'label'],index=None)
# Get first 5 images for both classes
malignant_samples = (train_data1[train_data1['label']==1]['image'].iloc[:5]).tolist()
benign_samples = (train_data1[train_data1['label']==0]['image'].iloc[:5]).tolist()
# Concat the data in a single list and del the above two list
samples = malignant_samples + benign_samples
del malignant_samples, benign_samples
# Plot the data
f, ax = plt.subplots(2,5, figsize=(30,10))
for i in range(10):
img = cv2.imread(samples[i])
ax[i//5, i%5].imshow(img, cmap='gray')
if i<5:
ax[i//5, i%5].set_title("benign")
else:
ax[i//5, i%5].set_title("malignant")
ax[i//5, i%5].axis('off')
ax[i//5, i%5].set_aspect('auto')
plt.show()
pipenv run python ./utils/sampleSet.py