Python SciKit-Learn Cheat Sheet
- Simple and efficient tools for predictive data analysis
- Accessible to everybody, and reusable in various contexts
- Built on NumPy, SciPy, and matplotlib
- Open source, commercially usable - BSD license
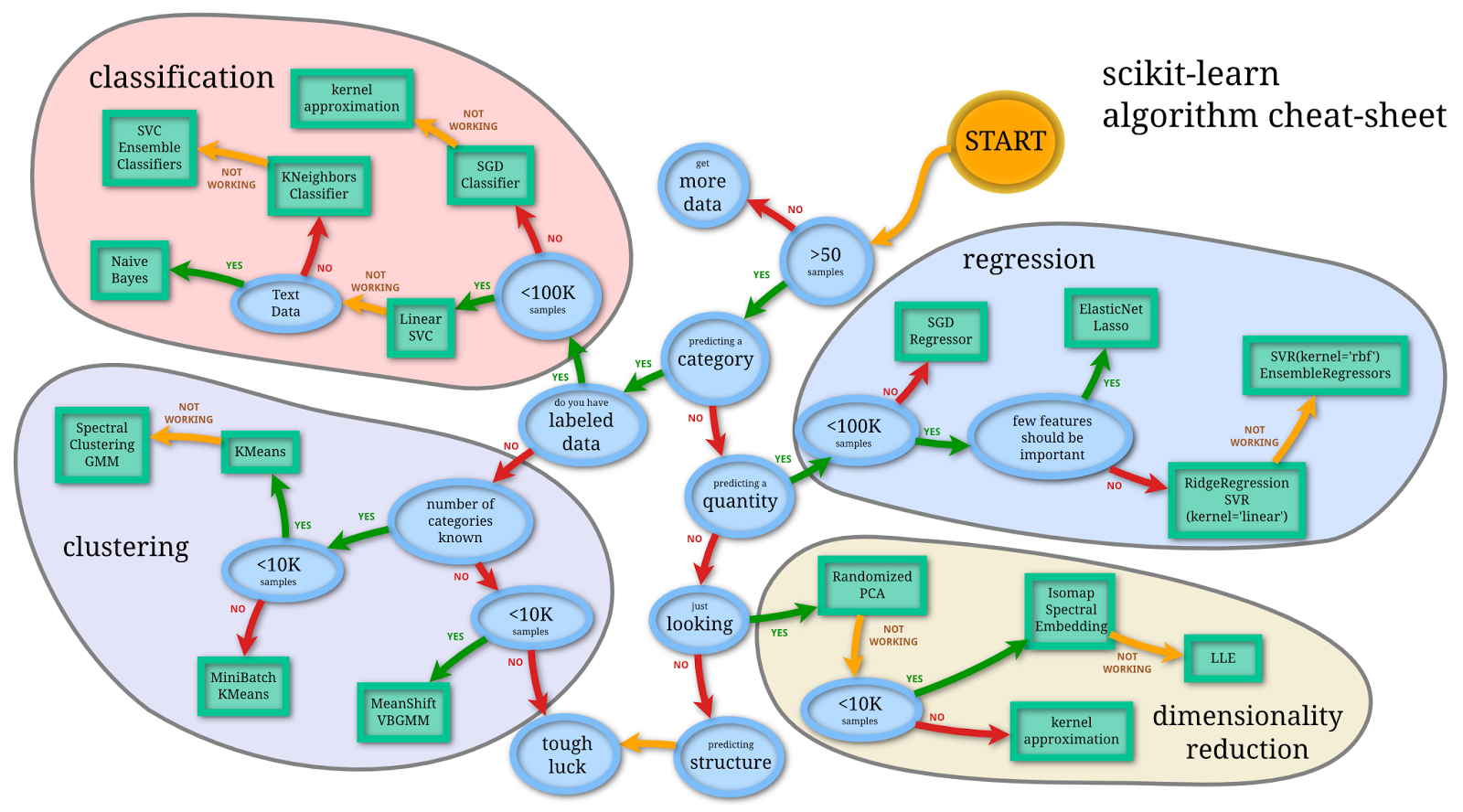
Image Source: SciKit Learn User Guide
Regressions ++ Classifications ++ Clustering ++ Dimensionality Reduction ++ Model Selection ++ Pre-processing
- Python SciKit-Learn Cheat Sheet
- Working with Missing Values
- Categorical Data Preprocessing
- Loading SK Datasets
- Supervised Learning - Regression Models
- Supervised Learning - Logistic Regression Model
- Supervised Learning - KNN Algorithm
- Supervised Learning - Decision Tree Classifier
- Supervised Learning - Random Forest Classifier
- Supervised Learning - SVC Model
- Supervised Learning - Boosting Methods
- Supervised Learning - Naive Bayes NLP
- Unsupervised Learning - KMeans Clustering
- Unsupervised Learning - Agglomerative Clustering
- Unsupervised Learning - Density-based Spatial Clustering (DBSCAN)
- Dimensionality Reduction - Principal Component Analysis (PCA)
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
from mpl_toolkits import mplot3d
import numpy as np
import pandas as pd
import plotly.express as px
from scipy.cluster import hierarchy
import seaborn as sns
from sklearn import svm
from sklearn.cluster import KMeans, AgglomerativeClustering, DBSCAN
from sklearn.datasets import load_iris, load_wine, fetch_20newsgroups, fetch_openml
from sklearn.impute import MissingIndicator, SimpleImputer
from sklearn.decomposition import PCA
from sklearn.ensemble import (
RandomForestClassifier,
RandomForestRegressor,
GradientBoostingRegressor,
AdaBoostRegressor,
GradientBoostingClassifier,
AdaBoostClassifier
)
from sklearn.feature_extraction.text import (
CountVectorizer,
TfidfTransformer,
TfidfVectorizer
)
from sklearn.linear_model import (
LinearRegression,
LogisticRegression,
Ridge,
ElasticNet
)
from sklearn.metrics import (
mean_absolute_error,
mean_squared_error,
classification_report,
confusion_matrix,
ConfusionMatrixDisplay,
accuracy_score
)
from sklearn.model_selection import (
train_test_split,
GridSearchCV,
cross_val_score,
cross_validate
)
from sklearn.naive_bayes import MultinomialNB
from sklearn.neighbors import KNeighborsClassifier, KNeighborsRegressor
from sklearn.pipeline import Pipeline, make_pipeline
from sklearn.preprocessing import (
MinMaxScaler,
StandardScaler,
OrdinalEncoder,
LabelEncoder,
OneHotEncoder,
PolynomialFeatures
)
from sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor
Working with Missing Values
X_missing = pd.DataFrame(
np.array([5,2,3,np.NaN,np.NaN,4,-3,2,1,8,np.NaN,4,10,np.NaN,5]).reshape(5,3)
)
X_missing.columns = ['f1','f2','f3']
X_missing
| f1 | f2 | f3 | |
|---|---|---|---|
| 0 | 5.0 | 2.0 | 3.0 |
| 1 | NaN | NaN | 4.0 |
| 2 | -3.0 | 2.0 | 1.0 |
| 3 | 8.0 | NaN | 4.0 |
| 4 | 10.0 | NaN | 5.0 |
X_missing.isnull().sum()
# f1 1
# f2 3
# f3 0
# dtype: int64
Missing Indicator
indicator = MissingIndicator(missing_values=np.NaN)
indicator = indicator.fit_transform(X_missing)
indicator = pd.DataFrame(indicator, columns=['a1', 'a2'])
indicator
| a1 | a2 | |
|---|---|---|
| 0 | False | False |
| 1 | True | True |
| 2 | False | False |
| 3 | False | True |
| 4 | False | True |
Simple Imputer
imputer_mean = SimpleImputer(missing_values=np.NaN, strategy='mean')
X_filled_mean = pd.DataFrame(imputer_mean.fit_transform(X_missing))
X_filled_mean.columns = ['f1','f2','f3']
X_filled_mean
| f1 | f2 | f3 | |
|---|---|---|---|
| 0 | 5.0 | 2.0 | 3.0 |
| 1 | 5.0 | 2.0 | 4.0 |
| 2 | -3.0 | 2.0 | 1.0 |
| 3 | 8.0 | 2.0 | 4.0 |
| 4 | 10.0 | 2.0 | 5.0 |
imputer_median = SimpleImputer(missing_values=np.NaN, strategy='median')
X_filled_median = pd.DataFrame(imputer_median.fit_transform(X_missing))
X_filled_median.columns = ['f1','f2','f3']
X_filled_median
| f1 | f2 | f3 | |
|---|---|---|---|
| 0 | 5.0 | 2.0 | 3.0 |
| 1 | 6.5 | 2.0 | 4.0 |
| 2 | -3.0 | 2.0 | 1.0 |
| 3 | 8.0 | 2.0 | 4.0 |
| 4 | 10.0 | 2.0 | 5.0 |
imputer_median = SimpleImputer(missing_values=np.NaN, strategy='most_frequent')
X_filled_median = pd.DataFrame(imputer_median.fit_transform(X_missing))
X_filled_median.columns = ['f1','f2','f3']
X_filled_median
| f1 | f2 | f3 | |
|---|---|---|---|
| 0 | 5.0 | 2.0 | 3.0 |
| 1 | -3.0 | 2.0 | 4.0 |
| 2 | -3.0 | 2.0 | 1.0 |
| 3 | 8.0 | 2.0 | 4.0 |
| 4 | 10.0 | 2.0 | 5.0 |
Drop Missing Data
X_missing_dropped = X_missing.dropna(axis=1)
X_missing_dropped
| f3 | |
|---|---|
| 0 | 3.0 |
| 1 | 4.0 |
| 2 | 1.0 |
| 3 | 4.0 |
| 4 | 5.0 |
X_missing_dropped = X_missing.dropna(axis=0).reset_index()
X_missing_dropped
| f1 | f2 | f3 | |
|---|---|---|---|
| 0 | 5.0 | 2.0 | 3.0 |
| 1 | -3.0 | 2.0 | 1.0 |
Categorical Data Preprocessing
X_cat_df = pd.DataFrame(
np.array([
['M', 'O-', 'medium'],
['M', 'O-', 'high'],
['F', 'O+', 'high'],
['F', 'AB', 'low'],
['F', 'B+', 'medium']
])
)
X_cat_df.columns = ['f1','f2','f3']
X_cat_df
| f1 | f2 | f3 | |
|---|---|---|---|
| 0 | M | O- | medium |
| 1 | M | O- | high |
| 2 | F | O+ | high |
| 3 | F | AB | low |
| 4 | F | B+ | medium |
Ordinal Encoder
encoder_ord = OrdinalEncoder(dtype='int')
X_cat_df.f3 = encoder_ord.fit_transform(X_cat_df.f3.values.reshape(-1, 1))
X_cat_df
| f1 | f2 | f3 | |
|---|---|---|---|
| 0 | M | O- | 2 |
| 1 | M | O- | 0 |
| 2 | F | O+ | 0 |
| 3 | F | AB | 1 |
| 4 | F | B+ | 2 |
Label Encoder
encoder_lab = LabelEncoder()
X_cat_df['f2'] = encoder_lab.fit_transform(X_cat_df['f2'])
X_cat_df
| f1 | f2 | f3 | |
|---|---|---|---|
| 0 | M | 3 | 2 |
| 1 | M | 3 | 0 |
| 2 | F | 2 | 0 |
| 3 | F | 0 | 1 |
| 4 | F | 1 | 2 |
OneHot Encoder
encoder_oh = OneHotEncoder(dtype='int')
onehot_df = pd.DataFrame(
encoder_oh.fit_transform(X_cat_df[['f1']])
.toarray(),
columns=['F', 'M']
)
onehot_df['f2'] = X_cat_df.f2
onehot_df['f3'] = X_cat_df.f3
onehot_df
| F | M | f2 | f3 | |
|---|---|---|---|---|
| 0 | 0 | 1 | 3 | 2 |
| 1 | 0 | 1 | 3 | 0 |
| 2 | 1 | 0 | 2 | 0 |
| 3 | 1 | 0 | 0 | 1 |
| 4 | 1 | 0 | 1 | 2 |
Loading SK Datasets
Toy Datasets
| load_iris(*[, return_X_y, as_frame]) | classification | Load and return the iris dataset. |
| load_diabetes(*[, return_X_y, as_frame, scaled]) | regression | Load and return the diabetes dataset. |
| load_digits(*[, n_class, return_X_y, as_frame]) | classification | Load and return the digits dataset. |
| load_linnerud(*[, return_X_y, as_frame]) | multi-output regression | Load and return the physical exercise Linnerud dataset. |
| load_wine(*[, return_X_y, as_frame]) | classification | Load and return the wine dataset. |
| load_breast_cancer(*[, return_X_y, as_frame]) | classification | Load and return the breast cancer wisconsin dataset. |
iris_ds = load_iris()
iris_data = iris_ds.data
col_names = iris_ds.feature_names
target_names = iris_ds.target_names
print(
'Iris Dataset',
'\n * Data array: ',
iris_data.shape,
'\n * Column names: ',
col_names,
'\n * Target names: ',
target_names
)
# Iris Dataset
# * Data array: (150, 4)
# * Column names: ['sepal length (cm)', 'sepal width (cm)', 'petal length (cm)', 'petal width (cm)']
# * Target names: ['setosa' 'versicolor' 'virginica']
iris_df = pd.DataFrame(data=iris_data, columns=col_names)
iris_df.head()
| sepal length (cm) | sepal width (cm) | petal length (cm) | petal width (cm) | |
|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 |
Real World Datasets
| fetch_olivetti_faces(*[, data_home, ...]) | classification | Load the Olivetti faces data-set from AT&T. |
| fetch_20newsgroups(*[, data_home, subset, ...]) | classification | Load the filenames and data from the 20 newsgroups dataset. |
| fetch_20newsgroups_vectorized(*[, subset, ...]) | classification | Load and vectorize the 20 newsgroups dataset. |
| fetch_lfw_people(*[, data_home, funneled, ...]) | classification | Load the Labeled Faces in the Wild (LFW) people dataset. |
| fetch_lfw_pairs(*[, subset, data_home, ...]) | classification | Load the Labeled Faces in the Wild (LFW) pairs dataset. |
| fetch_covtype(*[, data_home, ...]) | classification | Load the covertype dataset. |
| fetch_rcv1(*[, data_home, subset, ...]) | classification | Load the RCV1 multilabel dataset. |
| fetch_kddcup99(*[, subset, data_home, ...]) | classification | Load the kddcup99 dataset. |
| fetch_california_housing(*[, data_home, ...]) | regression | Load the California housing dataset. |
newsgroups_train = fetch_20newsgroups(subset='train')
train_data = newsgroups_train.data
col_names = newsgroups_train.filenames.shape
target_names = newsgroups_train.target.shape
print(
'Newsgroup - Train Subset',
'\n * Data array: ',
len(train_data),
'\n * Column names: ',
col_names,
'\n * Target names: ',
target_names
)
# Newsgroup - Train Subset
# * Data array: 11314
# * Column names: (11314,)
# * Target names: (11314,)
print('Target Names: ', newsgroups_train.target_names)
# Target Names: ['alt.atheism', 'comp.graphics', 'comp.os.ms-windows.misc', 'comp.sys.ibm.pc.hardware', 'comp.sys.mac.hardware', 'comp.windows.x', 'misc.forsale', 'rec.autos', 'rec.motorcycles', 'rec.sport.baseball', 'rec.sport.hockey', 'sci.crypt', 'sci.electronics', 'sci.med', 'sci.space', 'soc.religion.christian', 'talk.politics.guns', 'talk.politics.mideast', 'talk.politics.misc', 'talk.religion.misc']
OpenML Datasets
mice_ds = fetch_openml(name='miceprotein', version=4, parser="auto")
print(
'Mice Protein Dataset',
'\n * Data Shape: ',
mice_ds.data.shape,
'\n * Target Shape: ',
mice_ds.target.shape,
'\n * Target Names: ',
np.unique(mice_ds.target)
)
# Mice Protein Dataset
# * Data Shape: (1080, 77)
# * Target Shape: (1080,)
# * Target Names: ['c-CS-m' 'c-CS-s' 'c-SC-m' 'c-SC-s' 't-CS-m' 't-CS-s' 't-SC-m' 't-SC-s']
print(mice_ds.DESCR)
Supervised Learning - Regression Models
Simple Linear Regression
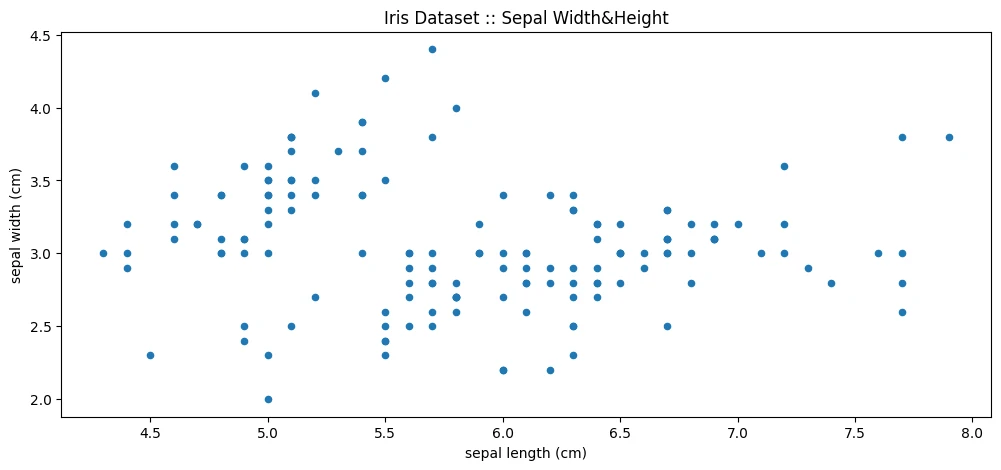
iris_df.plot(
figsize=(12,5),
kind='scatter',
x='sepal length (cm)',
y='sepal width (cm)',
title='Iris Dataset :: Sepal Width&Height'
)
print(iris_df.corr())
The Sepal Width has very little correlation to all other metrics but itself. While the other three correlate nicely:
| sepal length (cm) | sepal width (cm) | petal length (cm) | petal width (cm) | |
|---|---|---|---|---|
| sepal length (cm) | 1.000000 | -0.117570 | 0.871754 | 0.817941 |
| sepal width (cm) | -0.117570 | 1.000000 | -0.428440 | -0.366126 |
| petal length (cm) | 0.871754 | -0.428440 | 1.000000 | 0.962865 |
| petal width (cm) | 0.817941 | -0.366126 | 0.962865 | 1.000000 |
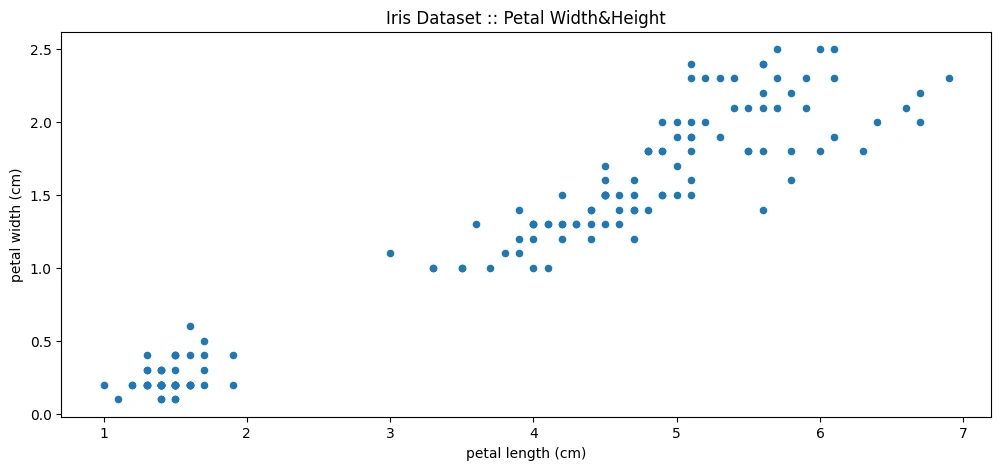
Data Pre-processing
iris_df['petal length (cm)'][:1]
# 0 1.4
# Name: petal length (cm), dtype: float64
iris_df['petal length (cm)'].values.reshape(-1,1)[:1]
# array([[1.4]])
# scikit expects a 2s imput => remove index
X = iris_df['petal length (cm)'].values.reshape(-1,1)
y = iris_df['petal width (cm)'].values.reshape(-1,1)
# train/test split
X_train, X_test, y_train, y_test = train_test_split(X,y,test_size=0.2)
print(X_train.shape, X_test.shape)
# (120, 1) (30, 1) 80:20 split
Model Training
regressor = LinearRegression()
regressor.fit(X_train,y_train)
intercept = regressor.intercept_
slope = regressor.coef_
print(' Intercept: ', intercept, '\n Slope: ', slope)
# Intercept: [-0.35135666]
# Correlation Coeficient: [[0.41310505]]
Predictions
y_pred = regressor.predict([X_test[0]])
print(' Prediction: ', y_pred, '\n True Value: ', y_test[0])
# Prediction: [[0.22699041]]
# True Value: [0.2]
def predict(value):
return (slope*value + intercept)[0][0]
print('Prediction: ', predict(X_test[0]))
# Prediction: [[0.22699041]]
iris_df['petal width (cm) prediction'] = iris_df['petal length (cm)'].apply(predict)
print(' Prediction: ', iris_df['petal width (cm) prediction'][0], '\n True Value: ', iris_df['petal width (cm)'][0])
# Prediction: 0.22699041280334376
# True Value: 0.2
iris_df.head(10)
| sepal length (cm) | sepal width (cm) | petal length (cm) | petal width (cm) | petal width (cm) prediction | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | 0.226990 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | 0.226990 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | 0.185680 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | 0.268301 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | 0.226990 |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | 0.350922 |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | 0.226990 |
| 7 | 5.0 | 3.4 | 1.5 | 0.2 | 0.268301 |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | 0.226990 |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | 0.268301 |
iris_df.plot(
figsize=(12,5),
kind='scatter',
x='petal width (cm)',
y='petal width (cm) prediction',
# no value in colorizing..just looks pretty
c='petal width (cm) prediction',
colormap='summer',
title='Iris Dataset - Sepal Width True vs Prediction'
)
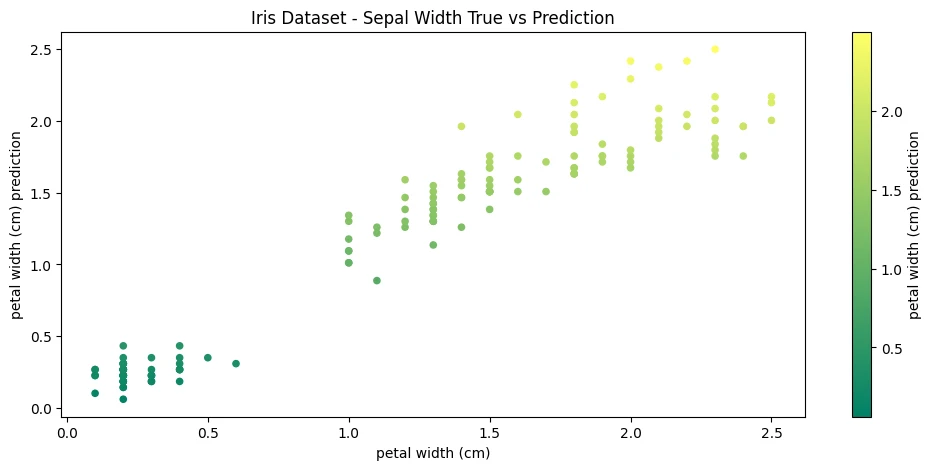
Model Evaluation
mae = mean_absolute_error(
iris_df['petal width (cm)'],
iris_df['petal width (cm) prediction']
)
mse = mean_squared_error(
iris_df['petal width (cm)'],
iris_df['petal width (cm) prediction']
)
rmse = np.sqrt(mse)
print(' MAE: ', mae, '\n MSE: ', mse, '\n RMSE: ', rmse)
# MAE: 0.1569441318761155
# MSE: 0.04209214667485277
# RMSE: 0.2051637070118708
ElasticNet Regression
Dataset
!wget https://raw.githubusercontent.com/Satish-Vennapu/DataScience/main/AMES_Final_DF.csv -P datasets
ames_df = pd.read_csv('datasets/AMES_Final_DF.csv')
ames_df.head(5).transpose()
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| Lot Frontage | 141.0 | 80.0 | 81.0 | 93.0 | 74.0 |
| Lot Area | 31770.0 | 11622.0 | 14267.0 | 11160.0 | 13830.0 |
| Overall Qual | 6.0 | 5.0 | 6.0 | 7.0 | 5.0 |
| Overall Cond | 5.0 | 6.0 | 6.0 | 5.0 | 5.0 |
| Year Built | 1960.0 | 1961.0 | 1958.0 | 1968.0 | 1997.0 |
| ... | |||||
| Sale Condition_AdjLand | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Sale Condition_Alloca | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Sale Condition_Family | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Sale Condition_Normal | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| Sale Condition_Partial | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 274 rows × 5 columns |
# the target value is:
ames_df['SalePrice']
| 0 | 215000 |
| 1 | 105000 |
| 2 | 172000 |
| 3 | 244000 |
| 4 | 189900 |
| ... | |
| 2920 | 142500 |
| 2921 | 131000 |
| 2922 | 132000 |
| 2923 | 170000 |
| 2924 | 188000 |
| Name: SalePrice, Length: 2925, dtype: int64 |
Preprocessing
# remove target column from training dataset
X_ames = ames_df.drop('SalePrice', axis=1)
y_ames = ames_df['SalePrice']
print(X_ames.shape, y_ames.shape)
# (2925, 273) (2925,)
# train/test split
X_ames_train, X_ames_test, y_ames_train, y_ames_test = train_test_split(
X_ames,
y_ames,
test_size=0.1,
random_state=101
)
print(X_ames_train.shape, X_ames_test.shape)
# (2632, 273) (293, 273)
# normalize feature set
scaler = StandardScaler()
X_ames_train_scaled = scaler.fit_transform(X_ames_train)
X_ames_test_scaled = scaler.transform(X_ames_test)
Grid Search for Hyperparameters
base_ames_elastic_net_model = ElasticNet(max_iter=int(1e4))
param_grid = \{
'alpha': [50, 75, 100, 125, 150],
'l1_ratio':[0.2, 0.4, 0.6, 0.8, 1.0]
\}
grid_ames_model = GridSearchCV(
estimator=base_ames_elastic_net_model,
param_grid=param_grid,
scoring='neg_mean_squared_error',
cv=5, verbose=1
)
grid_ames_model.fit(X_ames_train_scaled, y_ames_train)
print(
'Results:\nBest Estimator: ',
grid_ames_model.best_estimator_,
'\nBest Hyperparameter: ',
grid_ames_model.best_params_
)
Results:
- Best Estimator:
ElasticNet(alpha=125, l1_ratio=1.0, max_iter=10000) - Best Hyperparameter:
\{'alpha': 125, 'l1_ratio': 1.0\}
Model Evaluation
y_ames_pred = grid_ames_model.predict(X_ames_test_scaled)
print(
'MAE: ',
mean_absolute_error(y_ames_test, y_ames_pred),
'MSE: ',
mean_squared_error(y_ames_test, y_ames_pred),
'RMSE: ',
np.sqrt(mean_squared_error(y_ames_test, y_ames_pred))
)
# MAE: 14185.506207185055 MSE: 422714457.5190704 RMSE: 20560.020854052418
# average SalePrize
np.mean(ames_df['SalePrice'])
# 180815.53743589742
rel_error_avg = mean_absolute_error(y_ames_test, y_ames_pred) * 100 / np.mean(ames_df['SalePrice'])
print('Pridictions are on average off by: ', rel_error_avg.round(2), '%')
# Pridictions are on average off by: 7.85 %
plt.figure(figsize=(10,4))
plt.scatter(y_ames_test,y_ames_pred, c='mediumspringgreen', s=3)
plt.axline((0, 0), slope=1, color='dodgerblue', linestyle=(':'))
plt.title('Prediction Accuracy :: MAE:'+ str(mean_absolute_error(y_ames_test, y_ames_pred).round(2)) + 'US$')
plt.xlabel('True Sales Price')
plt.ylabel('Predicted Sales Price')
plt.savefig('assets/Scikit_Learn_11.webp', bbox_inches='tight')
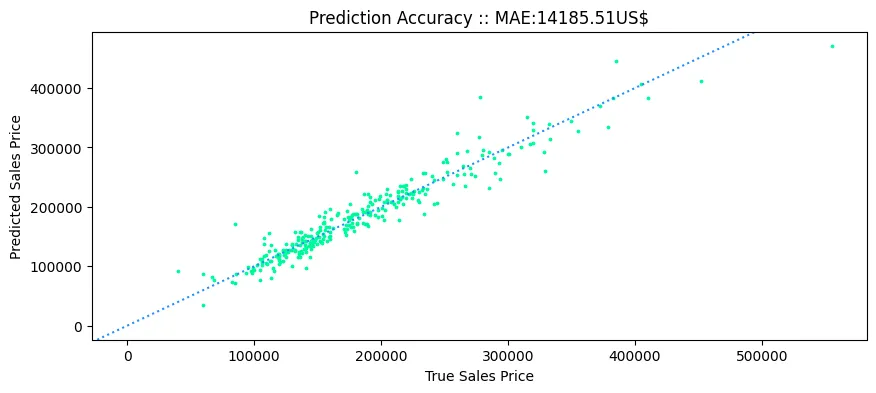
Multiple Linear Regression
Above I used the petal width and length to create a linear regression model. But as explored earlier we can also use the sepal length (only the sepal width does not show a linear correlation):
print(iris_df.corr())
| sepal length (cm) | sepal width (cm) | petal length (cm) | petal width (cm) | |
|---|---|---|---|---|
| sepal length (cm) | 1.000000 | -0.117570 | 0.871754 | 0.817941 |
| sepal width (cm) | -0.117570 | 1.000000 | -0.428440 | -0.366126 |
| petal length (cm) | 0.871754 | -0.428440 | 1.000000 | 0.962865 |
| petal width (cm) | 0.817941 | -0.366126 | 0.962865 | 1.000000 |
X_multi = iris_df[['petal length (cm)', 'sepal length (cm)']]
y = iris_df['petal width (cm)']
regressor_multi = LinearRegression()
regressor_multi.fit(X_multi, y)
intercept_multi = regressor_multi.intercept_
slope_multi = regressor_multi.coef_
print(' Intercept: ', intercept_multi, '\n Slope: ', slope_multi)
# Intercept: -0.00899597269816943
# Slope: [ 0.44937611 -0.08221782]
def predict_multi(petal_length, sepal_length):
return (slope_multi[0]*petal_length + slope_multi[1]*sepal_length + intercept_multi)
y_pred = predict_multi(
iris_df['petal length (cm)'][0],
iris_df['sepal length (cm)'][0]
)
print(' Prediction: ', y_pred, '\n True value: ', iris_df['petal width (cm)'][0])
# Prediction: 0.20081970121763193
# True value: 0.2
iris_df['petal width (cm) prediction (multi)'] = (
(
slope_multi[0] * iris_df['petal length (cm)']
) + (
slope_multi[1] * iris_df['sepal length (cm)']
) + (
intercept_multi
)
)
iris_df.head(10)
| sepal length (cm) | sepal width (cm) | petal length (cm) | petal width (cm) | petal width (cm) prediction | petal width (cm) prediction (multi) | |
|---|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | 0.226990 | 0.200820 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | 0.226990 | 0.217263 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | 0.185680 | 0.188769 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | 0.268301 | 0.286866 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | 0.226990 | 0.209041 |
| 5 | 5.4 | 3.9 | 1.7 | 0.4 | 0.350922 | 0.310967 |
| 6 | 4.6 | 3.4 | 1.4 | 0.3 | 0.226990 | 0.241929 |
| 7 | 5.0 | 3.4 | 1.5 | 0.2 | 0.268301 | 0.253979 |
| 8 | 4.4 | 2.9 | 1.4 | 0.2 | 0.226990 | 0.258372 |
| 9 | 4.9 | 3.1 | 1.5 | 0.1 | 0.268301 | 0.262201 |
iris_df.plot(
figsize=(12,5),
kind='scatter',
x='petal width (cm)',
y='petal width (cm) prediction (multi)',
c='petal width (cm) prediction',
colormap='summer',
title='Iris Dataset - Sepal Width True vs Prediction (multi)'
)
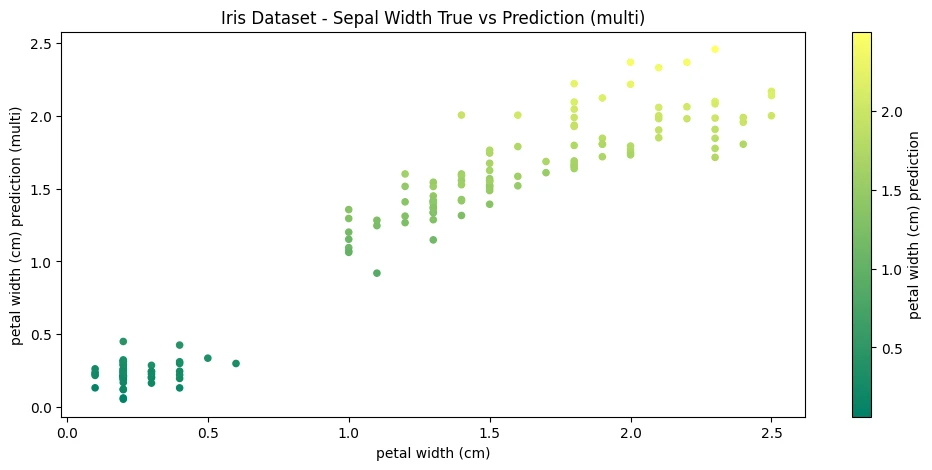
mae_multi = mean_absolute_error(
iris_df['petal width (cm)'],
iris_df['petal width (cm) prediction (multi)']
)
mse_multi = mean_squared_error(
iris_df['petal width (cm)'],
iris_df['petal width (cm) prediction (multi)']
)
rmse_multi = np.sqrt(mse_multi)
print(' MAE_Multi: ', mae_multi,' MAE: ', mae, '\n MSE_Multi: ', mse_multi, ' MSE: ', mse, '\n RMSE_Multi: ', rmse_multi, ' RMSE: ', rmse)
The accuracy of the model was improved by adding an additional, correlating value:
| Multi Regression | Single Regression | |
|---|---|---|
| Mean Absolute Error | 0.15562108079300102 | 0.1569441318761155 |
| Mean Squared Error | 0.04096208526408982 | 0.04209214667485277 |
| Root Mean Squared Error | 0.20239092189149646 | 0.2051637070118708 |
Supervised Learning - Logistic Regression Model
Binary Logistic Regression
Dataset
np.random.seed(666)
# generate 10 index values between 0-10
x_data_logistic_binary = np.random.randint(10, size=(10)).reshape(-1, 1)
# generate binary category for values above
y_data_logistic_binary = np.random.randint(2, size=10)
Model Fitting
logistic_binary_model = LogisticRegression(
solver='liblinear',
C=10.0,
random_state=0
)
logistic_binary_model.fit(x_data_logistic_binary, y_data_logistic_binary)
intercept_logistic_binary = logistic_binary_model.intercept_
slope_logistic_binary = logistic_binary_model.coef_
print(' Intercept: ', intercept_logistic_binary, '\n Slope: ', slope_logistic_binary)
# Intercept: [-0.4832956]
# Slope: [[0.11180522]]
Model Predictions
prob_pred_logistic_binary = logistic_binary_model.predict_proba(x_data_logistic_binary)
y_pred_logistic_binary = logistic_binary_model.predict(x_data_logistic_binary)
print('Prediction Probabilities: ', prob_pred[:1])
unique, counts = np.unique(y_pred_logistic_binary, return_counts=True)
print('Classes: ', unique, '| Number of Class Instances: ', counts)
# probabilities e.g. below -> 58% certainty that the first element is class 0
# Prediction Probabilities: [[0.58097284 0.41902716]]
# Classes: [0 1] | Number of Class Instances: [5 5]
Model Evaluation
conf_mtx = confusion_matrix(y_data_logistic_binary, y_pred_logistic_binary)
conf_mtx
# [2, 3] [TP, FP]
# [3, 2] [FN, TN]
report = classification_report(y_data_logistic_binary, y_pred_logistic_binary)
print(report)
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| 0 | 0.40 | 0.40 | 0.40 | 5 |
| 1 | 0.40 | 0.40 | 0.40 | 5 |
| accuracy | 0.40 | 10 | ||
| macro avg | 0.40 | 0.40 | 0.40 | 10 |
| weighted avg | 0.40 | 0.40 | 0.40 | 10 |
Logistic Regression Pipelines
Dataset Preprocessing
iris_ds = load_iris()
# train/test split
X_train_iris, X_test_iris, y_train_iris, y_test_iris = train_test_split(
iris_ds.data,
iris_ds.target,
test_size=0.2,
random_state=42
)
print(X_train_iris.shape, X_test_iris.shape)
# (120, 4) (30, 4)
Pipeline
pipe_iris = Pipeline([
('minmax', MinMaxScaler()),
('log_reg', LogisticRegression()),
])
pipe_iris.fit(X_train_iris, y_train_iris)
iris_score = pipe_iris.score(X_test_iris, y_test_iris)
print('Prediction Accuracy: ', iris_score.round(4)*100, '%')
# Prediction Accuracy: 96.67 %
Cross Validation
Train | Test Split
!wget https://raw.githubusercontent.com/reisanar/datasets/master/Advertising.csv -P datasets
adv_df = pd.read_csv('datasets/Advertising.csv')
adv_df.head(5)
| TV | Radio | Newspaper | Sales | |
|---|---|---|---|---|
| 0 | 230.1 | 37.8 | 69.2 | 22.1 |
| 1 | 44.5 | 39.3 | 45.1 | 10.4 |
| 2 | 17.2 | 45.9 | 69.3 | 9.3 |
| 3 | 151.5 | 41.3 | 58.5 | 18.5 |
| 4 | 180.8 | 10.8 | 58.4 | 12.9 |
# Split ds into features and targets
X_adv = adv_df.drop('Sales', axis=1)
y_adv = adv_df['Sales']
# 70:30 train/test split
X_adv_train, X_adv_test, y_adv_train, y_adv_test = train_test_split(
X_adv, y_adv, test_size=0.3, random_state=666
)
print(X_adv_train.shape, y_adv_train.shape)
# (140, 3) (140,)
# normalize features
scaler_adv = StandardScaler()
scaler_adv.fit(X_adv_train)
X_adv_train = scaler_adv.transform(X_adv_train)
X_adv_test = scaler_adv.transform(X_adv_test)
Model Fitting
model_adv1 = Ridge(
alpha=100.0
)
model_adv1.fit(X_adv_train, y_adv_train)
Model Evaluation
y_adv_pred = model_adv1.predict(X_adv_test)
mean_squared_error(y_adv_test, y_adv_pred)
# 6.528575771818745
Adjusting Hyper Parameter
model_adv2 = Ridge(
alpha=1.0
)
model_adv2.fit(X_adv_train, y_adv_train)
y_adv_pred2 = model_adv2.predict(X_adv_test)
mean_squared_error(y_adv_test, y_adv_pred2)
# 2.3319016551123535
Train | Validation | Test Split
# 70:30 train/temp split
X_adv_train, X_adv_temp, y_adv_train, y_adv_temp = train_test_split(
X_adv, y_adv, test_size=0.3, random_state=666
)
# 50:50 test/val split
X_adv_test, X_adv_val, y_adv_test, y_adv_val = train_test_split(
X_adv_temp, y_adv_temp, test_size=0.5, random_state=666
)
print(X_adv_train.shape, X_adv_test.shape, X_adv_val.shape)
# (140, 3) (30, 3) (30, 3)
# normalize features
scaler_adv = StandardScaler()
scaler_adv.fit(X_adv_train)
X_adv_train = scaler_adv.transform(X_adv_train)
X_adv_test = scaler_adv.transform(X_adv_test)
X_adv_val = scaler_adv.transform(X_adv_val)
Model Fitting and Evaluation
model_adv3 = Ridge(
alpha=100.0
)
model_adv3.fit(X_adv_train, y_adv_train)
# do evaluation with the validation set
y_adv_pred3 = model_adv3.predict(X_adv_val)
mean_squared_error(y_adv_val, y_adv_pred3)
# 7.136230975501291
Adjusting Hyper Parameter
model_adv4 = Ridge(
alpha=1.0
)
model_adv4.fit(X_adv_train, y_adv_train)
y_adv_pred4 = model_adv4.predict(X_adv_val)
mean_squared_error(y_adv_val, y_adv_pred4)
# 2.6393803874124435
# only once you are certain that you have the best performance
# do a final evaluation with the test set
y_adv4_final_pred = model_adv4.predict(X_adv_test)
mean_squared_error(y_adv_test, y_adv4_final_pred)
# 2.024422922812264
k-fold Cross Validation
Do a train/test split and segment the training set by k-folds (e.g. 5-10) and use each of those segments once to validate a training step. The resulting error is the average of all k errors.
Train-Test Split
# 70:30 train/temp split
X_adv_train, X_adv_test, y_adv_train, y_adv_test = train_test_split(
X_adv, y_adv, test_size=0.3, random_state=666
)
# normalize features
scaler_adv = StandardScaler()
scaler_adv.fit(X_adv_train)
X_adv_train = scaler_adv.transform(X_adv_train)
X_adv_test = scaler_adv.transform(X_adv_test)
Model Scoring
model_adv5 = Ridge(
alpha=100.0
)
# do a 5-fold cross-eval
scores = cross_val_score(
estimator=model_adv5,
X=X_adv_train,
y=y_adv_train,
scoring='neg_mean_squared_error',
cv=5
)
# take the mean of all five neg. error values
abs(scores.mean())
# 8.688107513529168
Adjusting Hyper Parameter
model_adv6 = Ridge(
alpha=1.0
)
# do a 5-fold cross-eval
scores = cross_val_score(
estimator=model_adv6,
X=X_adv_train,
y=y_adv_train,
scoring='neg_mean_squared_error',
cv=5
)
# take the mean of all five neg. error values
abs(scores.mean())
# 3.3419582340688576
Model Fitting and Final Evaluation
model_adv6.fit(X_adv_train, y_adv_train)
y_adv6_final_pred = model_adv6.predict(X_adv_test)
mean_squared_error(y_adv_test, y_adv6_final_pred)
# 2.3319016551123535
Cross Validate
Dataset (re-import)
adv_df = pd.read_csv('datasets/Advertising.csv')
X_adv = adv_df.drop('Sales', axis=1)
y_adv = adv_df['Sales']
# 70:30 train/test split
X_adv_train, X_adv_test, y_adv_train, y_adv_test = train_test_split(
X_adv, y_adv, test_size=0.3, random_state=666
)
# normalize features
scaler_adv = StandardScaler()
scaler_adv.fit(X_adv_train)
X_adv_train = scaler_adv.transform(X_adv_train)
X_adv_test = scaler_adv.transform(X_adv_test)
Model Scoring
model_adv7 = Ridge(
alpha=100.0
)
scores = cross_validate(
model_adv7,
X_adv_train,
y_adv_train,
scoring=[
'neg_mean_squared_error',
'neg_mean_absolute_error'
],
cv=10
)
scores_df = pd.DataFrame(scores)
scores_df
| fit_time | score_time | test_neg_mean_squared_error | test_neg_mean_absolute_error | |
|---|---|---|---|---|
| 0 | 0.016399 | 0.000749 | -12.539147 | -2.851864 |
| 1 | 0.000684 | 0.000452 | -2.806466 | -1.423516 |
| 2 | 0.000937 | 0.000782 | -11.142227 | -2.740332 |
| 3 | 0.001060 | 0.000633 | -7.237347 | -2.196963 |
| 4 | 0.001045 | 0.000738 | -11.313985 | -2.690813 |
| 5 | 0.000650 | 0.000510 | -3.169169 | -1.526568 |
| 6 | 0.000698 | 0.000429 | -6.578249 | -1.727616 |
| 7 | 0.000600 | 0.000423 | -5.740245 | -1.640964 |
| 8 | 0.000565 | 0.000463 | -10.268075 | -2.415688 |
| 9 | 0.000562 | 0.000487 | -10.641669 | -1.974407 |
abs(scores_df.mean())
| fit_time | 0.002320 |
| score_time | 0.000566 |
| test_neg_mean_squared_error | 8.143658 |
| test_neg_mean_absolute_error | 2.118873 |
| dtype: float64 |
Adjusting Hyper Parameter
model_adv8 = Ridge(
alpha=1.0
)
scores = cross_validate(
model_adv8,
X_adv_train,
y_adv_train,
scoring=[
'neg_mean_squared_error',
'neg_mean_absolute_error'
],
cv=10
)
abs(pd.DataFrame(scores).mean())
| fit_time | 0.001141 |
| score_time | 0.000777 |
| test_neg_mean_squared_error | 3.272673 |
| test_neg_mean_absolute_error | 1.345709 |
| dtype: float64 |
Model Fitting and Final Evaluation
model_adv8.fit(X_adv_train, y_adv_train)
y_adv8_final_pred = model_adv8.predict(X_adv_test)
mean_squared_error(y_adv_test, y_adv8_final_pred)
# 2.3319016551123535
Grid Search
Loop through a set of hyperparameters to find an optimum.
Hyperparameter Search
base_elastic_net_model = ElasticNet()
param_grid = \{
'alpha': [0.1, 1, 5, 10, 50, 100],
'l1_ratio':[0.1, 0.3, 0.5, 0.7, 0.9, 1.0]
\}
grid_model = GridSearchCV(
estimator=base_elastic_net_model,
param_grid=param_grid,
scoring='neg_mean_squared_error',
cv=5, verbose=2
)
grid_model.fit(X_adv_train, y_adv_train)
print(
'Results:\nBest Estimator: ',
grid_model.best_estimator_,
'\nBest Hyperparameter: ',
grid_model.best_params_
)
Results:
- Best Estimator:
ElasticNet(alpha=0.1, l1_ratio=1.0) - Best Hyperparameter:
\{'alpha': 0.1, 'l1_ratio': 1.0\}
gridcv_results = pd.DataFrame(grid_model.cv_results_)
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_alpha | param_l1_ratio | params | split0_test_score | split1_test_score | split2_test_score | split3_test_score | split4_test_score | mean_test_score | std_test_score | rank_test_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.001156 | 0.000160 | 0.000449 | 0.000038 | 0.1 | 0.1 | {'alpha': 0.1, 'l1_ratio': 0.1} | -1.924119 | -3.384152 | -3.588444 | -3.703040 | -5.091974 | -3.538346 | 1.007264 | 6 |
| 1 | 0.001144 | 0.000181 | 0.000407 | 0.000091 | 0.1 | 0.3 | {'alpha': 0.1, 'l1_ratio': 0.3} | -1.867117 | -3.304382 | -3.561106 | -3.623188 | -5.061781 | -3.483515 | 1.016000 | 5 |
| 2 | 0.000623 | 0.000026 | 0.000272 | 0.000052 | 0.1 | 0.5 | {'alpha': 0.1, 'l1_ratio': 0.5} | -1.812633 | -3.220727 | -3.539711 | -3.547572 | -5.043259 | -3.432780 | 1.028406 | 4 |
| 3 | 0.000932 | 0.000165 | 0.000321 | 0.000060 | 0.1 | 0.7 | {'alpha': 0.1, 'l1_ratio': 0.7} | -1.750153 | -3.144120 | -3.525226 | -3.477228 | -5.034008 | -3.386147 | 1.046722 | 3 |
| 4 | 0.000725 | 0.000106 | 0.000259 | 0.000024 | 0.1 | 0.9 | {'alpha': 0.1, 'l1_ratio': 0.9} | -1.693440 | -3.075686 | -3.518777 | -3.413393 | -5.029683 | -3.346196 | 1.065195 | 2 |
| 5 | 0.000654 | 0.000053 | 0.000274 | 0.000026 | 0.1 | 1.0 | {'alpha': 0.1, 'l1_ratio': 1.0} | -1.667506 | -3.044928 | -3.518866 | -3.384363 | -5.031297 | -3.329392 | 1.075006 | 1 |
| 6 | 0.000595 | 0.000016 | 0.000244 | 0.000002 | 1 | 0.1 | {'alpha': 1, 'l1_ratio': 0.1} | -8.575470 | -11.021534 | -8.212152 | -6.808719 | -10.792072 | -9.081990 | 1.604192 | 12 |
| 7 | 0.000591 | 0.000018 | 0.000244 | 0.000002 | 1 | 0.3 | {'alpha': 1, 'l1_ratio': 0.3} | -8.131855 | -10.448423 | -7.774620 | -6.179358 | -10.071728 | -8.521197 | 1.569173 | 11 |
| 8 | 0.000628 | 0.000049 | 0.000266 | 0.000023 | 1 | 0.5 | {'alpha': 1, 'l1_ratio': 0.5} | -7.519809 | -9.562473 | -7.261824 | -5.453399 | -9.213320 | -7.802165 | 1.481785 | 10 |
| 9 | 0.000594 | 0.000015 | 0.000243 | 0.000002 | 1 | 0.7 | {'alpha': 1, 'l1_ratio': 0.7} | -6.614835 | -8.351711 | -6.702104 | -4.698977 | -8.230616 | -6.919649 | 1.329741 | 9 |
| 10 | 0.000714 | 0.000108 | 0.000268 | 0.000033 | 1 | 0.9 | {'alpha': 1, 'l1_ratio': 0.9} | -5.537250 | -6.887828 | -6.148400 | -4.106124 | -7.101573 | -5.956235 | 1.078430 | 8 |
| 11 | 0.000649 | 0.000067 | 0.000263 | 0.000028 | 1 | 1.0 | {'alpha': 1, 'l1_ratio': 1.0} | -4.932027 | -6.058207 | -5.892529 | -3.798441 | -6.472871 | -5.430815 | 0.959804 | 7 |
| 12 | 0.000645 | 0.000042 | 0.000264 | 0.000040 | 5 | 0.1 | {'alpha': 5, 'l1_ratio': 0.1} | -21.863798 | -25.767488 | -18.768865 | -12.608680 | -23.207907 | -20.443347 | 4.520904 | 13 |
| 13 | 0.000617 | 0.000030 | 0.000281 | 0.000038 | 5 | 0.3 | {'alpha': 5, 'l1_ratio': 0.3} | -23.626694 | -27.439028 | -20.266203 | -12.788078 | -24.609195 | -21.745840 | 5.031493 | 14 |
| 14 | 0.000599 | 0.000011 | 0.000249 | 0.000013 | 5 | 0.5 | {'alpha': 5, 'l1_ratio': 0.5} | -26.202964 | -29.867138 | -22.527913 | -13.423857 | -26.835934 | -23.771561 | 5.675911 | 15 |
| 15 | 0.000588 | 0.000013 | 0.000276 | 0.000035 | 5 | 0.7 | {'alpha': 5, 'l1_ratio': 0.7} | -27.768946 | -33.428462 | -23.506474 | -14.599984 | -29.112276 | -25.683228 | 6.382379 | 17 |
| 16 | 0.000580 | 0.000003 | 0.000271 | 0.000001 | 5 | 0.9 | {'alpha': 5, 'l1_ratio': 0.9} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 17 | 0.000591 | 0.000011 | 0.000259 | 0.000021 | 5 | 1.0 | {'alpha': 5, 'l1_ratio': 1.0} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 18 | 0.000632 | 0.000028 | 0.000250 | 0.000012 | 10 | 0.1 | {'alpha': 10, 'l1_ratio': 0.1} | -26.179546 | -30.396420 | -22.386698 | -14.596498 | -27.292337 | -24.170300 | 5.429322 | 16 |
| 19 | 0.000593 | 0.000020 | 0.000239 | 0.000001 | 10 | 0.3 | {'alpha': 10, 'l1_ratio': 0.3} | -28.704426 | -33.379967 | -24.561645 | -15.634153 | -29.883725 | -26.432783 | 6.090062 | 18 |
| 20 | 0.000595 | 0.000036 | 0.000245 | 0.000013 | 10 | 0.5 | {'alpha': 10, 'l1_ratio': 0.5} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 21 | 0.000610 | 0.000053 | 0.000258 | 0.000015 | 10 | 0.7 | {'alpha': 10, 'l1_ratio': 0.7} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 22 | 0.000597 | 0.000022 | 0.000248 | 0.000015 | 10 | 0.9 | {'alpha': 10, 'l1_ratio': 0.9} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 23 | 0.000623 | 0.000057 | 0.000305 | 0.000076 | 10 | 1.0 | {'alpha': 10, 'l1_ratio': 1.0} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 24 | 0.000602 | 0.000016 | 0.000252 | 0.000013 | 50 | 0.1 | {'alpha': 50, 'l1_ratio': 0.1} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 25 | 0.000577 | 0.000009 | 0.000238 | 0.000001 | 50 | 0.3 | {'alpha': 50, 'l1_ratio': 0.3} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 26 | 0.000607 | 0.000046 | 0.000245 | 0.000010 | 50 | 0.5 | {'alpha': 50, 'l1_ratio': 0.5} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 27 | 0.000569 | 0.000004 | 0.000259 | 0.000012 | 50 | 0.7 | {'alpha': 50, 'l1_ratio': 0.7} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 28 | 0.000582 | 0.000022 | 0.000244 | 0.000011 | 50 | 0.9 | {'alpha': 50, 'l1_ratio': 0.9} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 29 | 0.000603 | 0.000041 | 0.000251 | 0.000015 | 50 | 1.0 | {'alpha': 50, 'l1_ratio': 1.0} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 30 | 0.000670 | 0.000106 | 0.000251 | 0.000013 | 100 | 0.1 | {'alpha': 100, 'l1_ratio': 0.1} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 31 | 0.000764 | 0.000179 | 0.000343 | 0.000054 | 100 | 0.3 | {'alpha': 100, 'l1_ratio': 0.3} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 32 | 0.000623 | 0.000077 | 0.000244 | 0.000007 | 100 | 0.5 | {'alpha': 100, 'l1_ratio': 0.5} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 33 | 0.000817 | 0.000156 | 0.000329 | 0.000076 | 100 | 0.7 | {'alpha': 100, 'l1_ratio': 0.7} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 34 | 0.000590 | 0.000017 | 0.000242 | 0.000004 | 100 | 0.9 | {'alpha': 100, 'l1_ratio': 0.9} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
| 35 | 0.000595 | 0.000027 | 0.000242 | 0.000007 | 100 | 1.0 | {'alpha': 100, 'l1_ratio': 1.0} | -29.868949 | -34.423737 | -25.623955 | -16.750237 | -31.056181 | -27.544612 | 6.087093 | 19 |
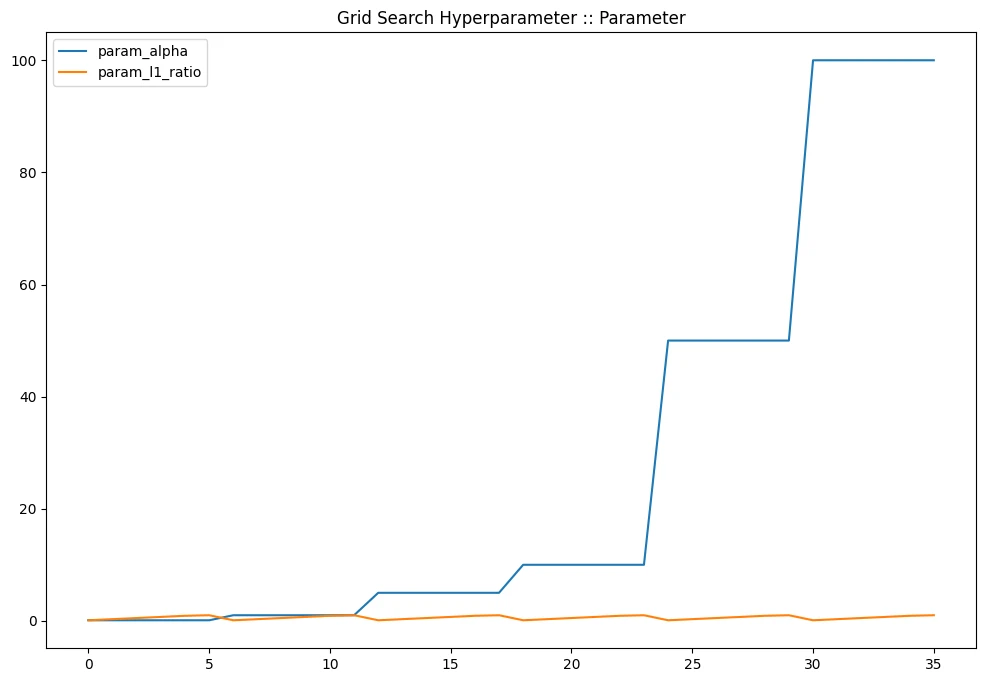
gridcv_results[
[
'param_alpha',
'param_l1_ratio'
]
].plot(title='Grid Search Hyperparameter :: Parameter', figsize=(12,8))
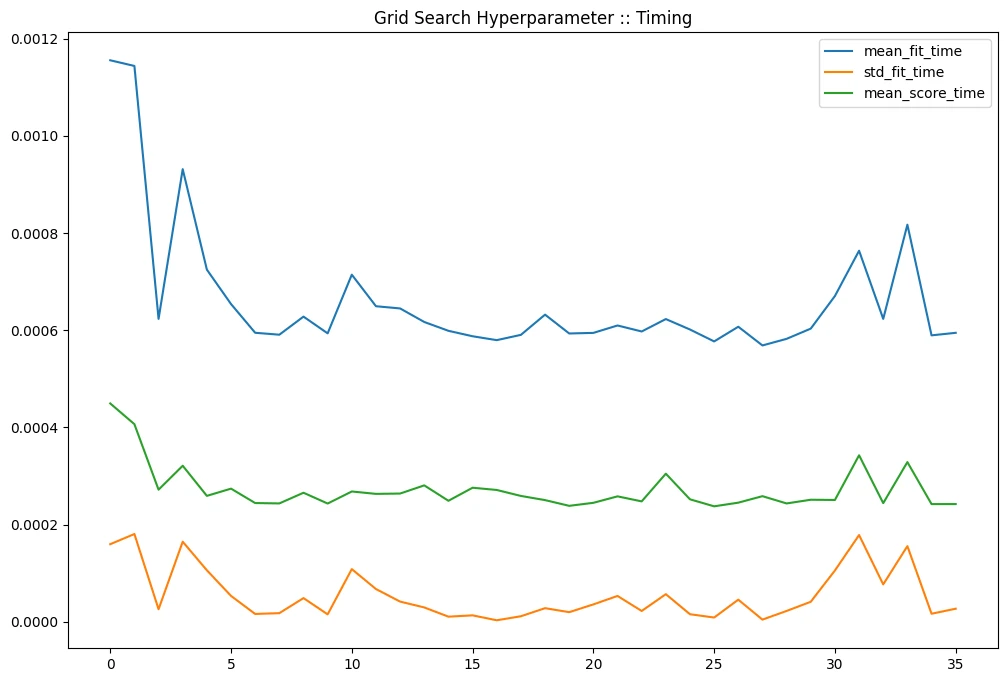
gridcv_results[
[
'mean_fit_time',
'std_fit_time',
'mean_score_time'
]
].plot(title='Grid Search Hyperparameter :: Timing', figsize=(12,8))
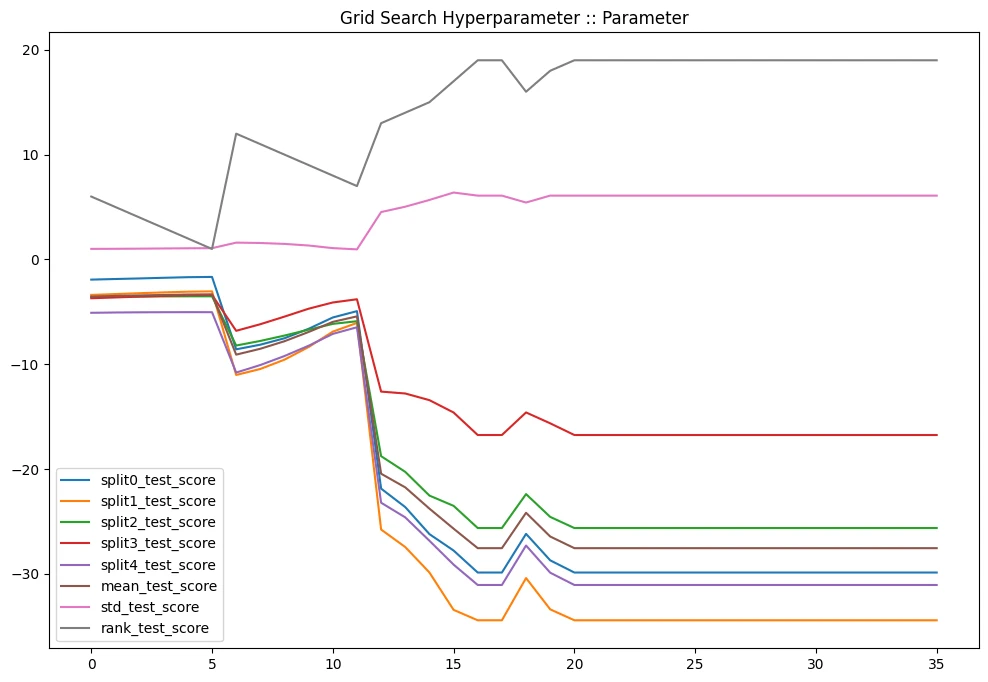
gridcv_results[
[
'split0_test_score',
'split1_test_score',
'split2_test_score',
'split3_test_score',
'split4_test_score',
'mean_test_score',
'std_test_score',
'rank_test_score'
]
].plot(title='Grid Search Hyperparameter :: Parameter', figsize=(12,8))
Model Evaluation
y_grid_pred = grid_model.predict(X_adv_test)
mean_squared_error(y_adv_test, y_grid_pred)
# 2.380865536033581
Supervised Learning - KNN Algorithm
Dataset
wine = load_wine()
print(wine.data.shape)
print(wine.feature_names)
print(wine.data[:1])
# (178, 13)
# ['alcohol', 'malic_acid', 'ash', 'alcalinity_of_ash', 'magnesium', 'total_phenols', 'flavanoids', 'nonflavanoid_phenols', 'proanthocyanins', 'color_intensity', 'hue', 'od280/od315_of_diluted_wines', 'proline']
# [[1.423e+01 1.710e+00 2.430e+00 1.560e+01 1.270e+02 2.800e+00 3.060e+00
# 2.800e-01 2.290e+00 5.640e+00 1.040e+00 3.920e+00 1.065e+03]]
wine_df = pd.DataFrame(data=wine.data, columns=wine.feature_names)
wine_df.head(2).T
| 0 | 1 | |
|---|---|---|
| alcohol | 14.23 | 13.20 |
| malic_acid | 1.71 | 1.78 |
| ash | 2.43 | 2.14 |
| alcalinity_of_ash | 15.60 | 11.20 |
| magnesium | 127.00 | 100.00 |
| total_phenols | 2.80 | 2.65 |
| flavanoids | 3.06 | 2.76 |
| nonflavanoid_phenols | 0.28 | 0.26 |
| proanthocyanins | 2.29 | 1.28 |
| color_intensity | 5.64 | 4.38 |
| hue | 1.04 | 1.05 |
| od280/od315_of_diluted_wines | 3.92 | 3.40 |
| proline | 1065.00 | 1050.00 |
Data Pre-processing
# normalization
scaler = MinMaxScaler()
scaler.fit(wine.data)
wine_norm = scaler.fit_transform(wine.data)
# train/test split
X_train_wine, X_test_wine, y_train_wine, y_test_wine = train_test_split(
wine_norm,
wine.target,
test_size=0.3
)
print(X_train_wine.shape, X_test_wine.shape)
# (124, 13) (54, 13)
Model Fitting
# model for k=3
knn = KNeighborsClassifier(n_neighbors=3)
knn.fit(X_train_wine, y_train_wine)
y_pred_wine_knn3 = knn.predict(X_test_wine)
print('Accuracy Score: ', (accuracy_score(y_test_wine, y_pred_wine_knn3)*100).round(2), '%')
# Accuracy Score: 98.15 %
# model for k=5
knn = KNeighborsClassifier(n_neighbors=5)
knn.fit(X_train_wine, y_train_wine)
y_pred_wine_knn5 = knn.predict(X_test_wine)
print('Accuracy Score: ', (accuracy_score(y_test_wine, y_pred_wine_knn5)*100).round(2), '%')
# Accuracy Score: 98.15 %
# model for k=7
knn = KNeighborsClassifier(n_neighbors=7)
knn.fit(X_train_wine, y_train_wine)
y_pred_wine_knn7 = knn.predict(X_test_wine)
print('Accuracy Score: ', (accuracy_score(y_test_wine, y_pred_wine_knn7)*100).round(2), '%')
# Accuracy Score: 96.3 %
# model for k=9
knn = KNeighborsClassifier(n_neighbors=7)
knn.fit(X_train_wine, y_train_wine)
y_pred_wine_knn7 = knn.predict(X_test_wine)
print('Accuracy Score: ', (accuracy_score(y_test_wine, y_pred_wine_knn7)*100).round(2), '%')
# Accuracy Score: 96.3 %
Supervised Learning - Decision Tree Classifier
- Does not require normalization
- Is not sensitive to missing values
Dataset
!wget https://gist.githubusercontent.com/Dviejopomata/ea5869ba4dcff84f8c294dc7402cd4a9/raw/4671f90b8b04ba4db9d67acafaa4c0827cd233c2/bill_authentication.csv -P datasets
bill_auth_df = pd.read_csv('datasets/bill_authentication.csv')
bill_auth_df.head(3)
| Variance | Skewness | Curtosis | Entropy | Class | |
|---|---|---|---|---|---|
| 0 | 3.6216 | 8.6661 | -2.8073 | -0.44699 | 0 |
| 1 | 4.5459 | 8.1674 | -2.4586 | -1.46210 | 0 |
| 2 | 3.8660 | -2.6383 | 1.9242 | 0.10645 | 0 |
Preprocessing
# remove target feature from training set
X_bill = bill_auth_df.drop('Class', axis=1)
y_bill = bill_auth_df['Class']
X_train_bill, X_test_bill, y_train_bill, y_test_bill = train_test_split(X_bill, y_bill, test_size=0.2)
Model Fitting
tree_classifier = DecisionTreeClassifier()
tree_classifier.fit(X_train_bill, y_train_bill)
Evaluation
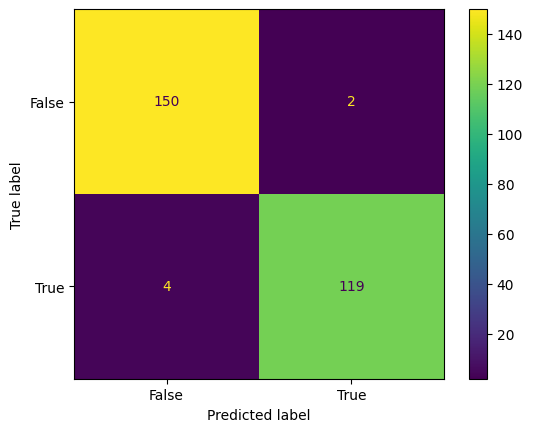
y_pred_bill = tree_classifier.predict(X_test_bill)
conf_mtx_bill = confusion_matrix(y_test_bill, y_pred_bill)
conf_mtx_bill
# array([[150, 2],
# [ 4, 119]])
conf_mtx_bill_plot = ConfusionMatrixDisplay(
confusion_matrix=conf_mtx_bill,
display_labels=[False,True]
)
conf_mtx_bill_plot.plot()
plt.show()
report_bill = classification_report(
y_test_bill, y_pred_bill
)
print(report_bill)
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| 0 | 0.97 | 0.99 | 0.98 | 152 |
| 1 | 0.98 | 0.97 | 0.98 | 123 |
| accuracy | 0.98 | 275 | ||
| macro avg | 0.98 | 0.98 | 0.98 | 275 |
| weighted avg | 0.98 | 0.98 | 0.98 | 275 |
Supervised Learning - Random Forest Classifier
- Does not require normalization
- Is not sensitive to missing values
- Low risk of overfitting
- Efficient with large datasets
- High accuracy
Dataset
!wget https://raw.githubusercontent.com/xjcjiacheng/data-analysis/master/heart%20disease%20UCI/heart.csv -P datasets
heart_df = pd.read_csv('datasets/heart.csv')
heart_df.head(5)
| age | sex | cp | trestbps | chol | fbs | restecg | thalach | exang | oldpeak | slope | ca | thal | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 63 | 1 | 3 | 145 | 233 | 1 | 0 | 150 | 0 | 2.3 | 0 | 0 | 1 | 1 |
| 1 | 37 | 1 | 2 | 130 | 250 | 0 | 1 | 187 | 0 | 3.5 | 0 | 0 | 2 | 1 |
| 2 | 41 | 0 | 1 | 130 | 204 | 0 | 0 | 172 | 0 | 1.4 | 2 | 0 | 2 | 1 |
| 3 | 56 | 1 | 1 | 120 | 236 | 0 | 1 | 178 | 0 | 0.8 | 2 | 0 | 2 | 1 |
| 4 | 57 | 0 | 0 | 120 | 354 | 0 | 1 | 163 | 1 | 0.6 | 2 | 0 | 2 | 1 |
Preprocessing
# remove target feature from training set
X_heart = heart_df.drop('target', axis=1)
y_heart = heart_df['target']
X_train_heart, X_test_heart, y_train_heart, y_test_heart = train_test_split(
X_heart,
y_heart,
test_size=0.2,
random_state=0
)
Model Fitting
forest_classifier = RandomForestClassifier(n_estimators=10, criterion='entropy')
forest_classifier.fit(X_train_heart, y_train_heart)
Evaluation
y_pred_heart = forest_classifier.predict(X_test_heart)
conf_mtx_heart = confusion_matrix(y_test_heart, y_pred_heart)
conf_mtx_heart
# array([[24, 3],
# [ 5, 29]])
conf_mtx_heart_plot = ConfusionMatrixDisplay(
confusion_matrix=conf_mtx_heart,
display_labels=[False,True]
)
conf_mtx_heart_plot.plot()
plt.show()
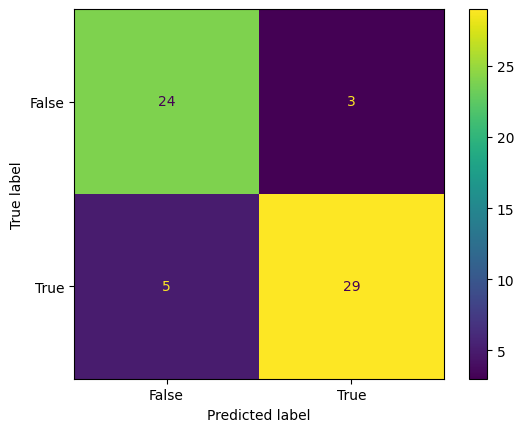
report_heart = classification_report(
y_test_heart, y_pred_heart
)
print(report_heart)
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| 0 | 0.83 | 0.89 | 0.86 | 27 |
| 1 | 0.91 | 0.85 | 0.88 | 34 |
| accuracy | 0.87 | 61 | ||
| macro avg | 0.87 | 0.87 | 0.87 | 61 |
| weighted avg | 0.87 | 0.87 | 0.87 | 61 |
Random Forest Hyperparameter Tuning
Testing Hyperparameters
rdnfor_classifier = RandomForestClassifier(
n_estimators=2,
min_samples_split=2,
min_samples_leaf=1,
criterion='entropy'
)
rdnfor_classifier.fit(X_train_heart, y_train_heart)
rdnfor_pred = rdnfor_classifier.predict(X_test_heart)
print('Accuracy Score: ', accuracy_score(y_test_heart, rdnfor_pred).round(4)*100, '%')
# Accuracy Score: 73.77 %
Grid-Search Cross-Validation
Try a set of values for selected Hyperparameter to find the optimal configuration.
param_grid = \{
'n_estimators': [5, 25, 50, 75,100, 125],
'min_samples_split': [1,2,3],
'min_samples_leaf': [1,2,3],
'criterion': ['gini', 'entropy', 'log_loss'],
'max_features' : ['sqrt', 'log2']
\}
grid_search = GridSearchCV(
estimator = rdnfor_classifier,
param_grid = param_grid
)
grid_search.fit(X_train_heart, y_train_heart)
print('Best Parameter: ', grid_search.best_params_)
# Best Parameter: \{
# 'criterion': 'entropy',
# 'max_features': 'sqrt',
# 'min_samples_leaf': 2,
# 'min_samples_split': 1,
# 'n_estimators': 25
# \}
rdnfor_classifier_optimized = RandomForestClassifier(
n_estimators=25,
min_samples_split=1,
min_samples_leaf=2,
criterion='entropy',
max_features='sqrt'
)
rdnfor_classifier_optimized.fit(X_train_heart, y_train_heart)
rdnfor_pred_optimized = rdnfor_classifier_optimized.predict(X_test_heart)
print('Accuracy Score: ', accuracy_score(y_test_heart, rdnfor_pred_optimized).round(4)*100, '%')
# Accuracy Score: 85.25 %
Random Forest Classifier 1 - Penguins
!wget https://github.com/remijul/dataset/raw/master/penguins_size.csv -P datasets
peng_df = pd.read_csv('datasets/penguins_size.csv')
peng_df = peng_df.dropna()
peng_df.head(5)
| species | island | culmen_length_mm | culmen_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | MALE |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | FEMALE |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | FEMALE |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | FEMALE |
| 5 | Adelie | Torgersen | 39.3 | 20.6 | 190.0 | 3650.0 | MALE |
# drop labels and encode string values
X_peng = pd.get_dummies(peng_df.drop('species', axis=1),drop_first=True)
y_peng = peng_df['species']
# train/test split
X_peng_train, X_peng_test, y_peng_train, y_peng_test = train_test_split(
X_peng,
y_peng,
test_size=0.3,
random_state=42
)
# creating the model
rfc_peng = RandomForestClassifier(
n_estimators=10,
max_features='sqrt',
random_state=42
)
# model training and running predictions
rfc_peng.fit(X_peng_train, y_peng_train)
peng_pred = rfc_peng.predict(X_peng_test)
print('Accuracy Score: ',accuracy_score(y_peng_test, peng_pred, normalize=True).round(4)*100, '%')
# Accuracy Score: 98.02 %
Feature Importance
# feature importance for classification
peng_index = ['importance']
peng_data_columns = pd.Series(X_peng.columns)
peng_importance_array = rfc_peng.feature_importances_
peng_importance_df = pd.DataFrame(peng_importance_array, peng_data_columns, peng_index)
peng_importance_df
| importance | |
|---|---|
| culmen_length_mm | 0.288928 |
| culmen_depth_mm | 0.111021 |
| flipper_length_mm | 0.357994 |
| body_mass_g | 0.025477 |
| island_Dream | 0.178498 |
| island_Torgersen | 0.031042 |
| sex_FEMALE | 0.004716 |
| sex_MALE | 0.002324 |
peng_importance_df.sort_values(
by='importance',
ascending=False
).plot(
kind='barh',
title='Feature Importance for Species Classification',
figsize=(12,4)
)
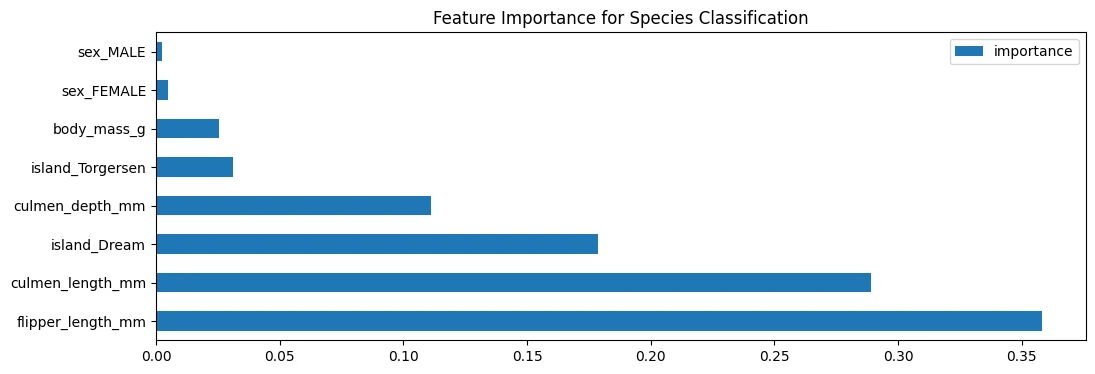
Model Evaluation
report_peng = classification_report(y_peng_test, peng_pred)
print(report_peng)
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| Adelie | 0.98 | 0.98 | 0.98 | 49 |
| Chinstrap | 0.94 | 0.94 | 0.94 | 18 |
| Gentoo | 1.00 | 1.00 | 1.00 | 34 |
| accuracy | 0.98 | 101 | ||
| macro avg | 0.97 | 0.97 | 0.97 | 101 |
| weighted avg | 0.98 | 0.98 | 0.98 | 101 |
conf_mtx_peng = confusion_matrix(y_peng_test, peng_pred)
conf_mtx_peng_plot = ConfusionMatrixDisplay(
confusion_matrix=conf_mtx_peng
)
conf_mtx_peng_plot.plot(cmap='plasma')
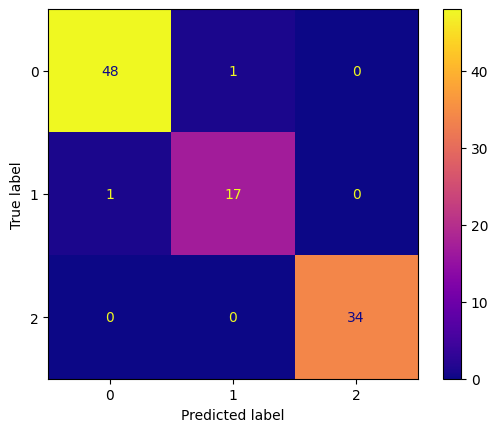
Random Forest Classifier - Banknote Authentication
!wget https://github.com/jbrownlee/Datasets/raw/master/banknote_authentication.csv -P datasets
money_df = pd.read_csv('datasets/data-banknote-authentication.csv')
money_df.head(5)
| Variance_Wavelet | Skewness_Wavelet | Curtosis_Wavelet | Image_Entropy | Class | |
|---|---|---|---|---|---|
| 0 | 3.62160 | 8.6661 | -2.8073 | -0.44699 | 0 |
| 1 | 4.54590 | 8.1674 | -2.4586 | -1.46210 | 0 |
| 2 | 3.86600 | -2.6383 | 1.9242 | 0.10645 | 0 |
| 3 | 3.45660 | 9.5228 | -4.0112 | -3.59440 | 0 |
| 4 | 0.32924 | -4.4552 | 4.5718 | -0.98880 | 0 |
sns.pairplot(money_df, hue='Class', palette='winter')
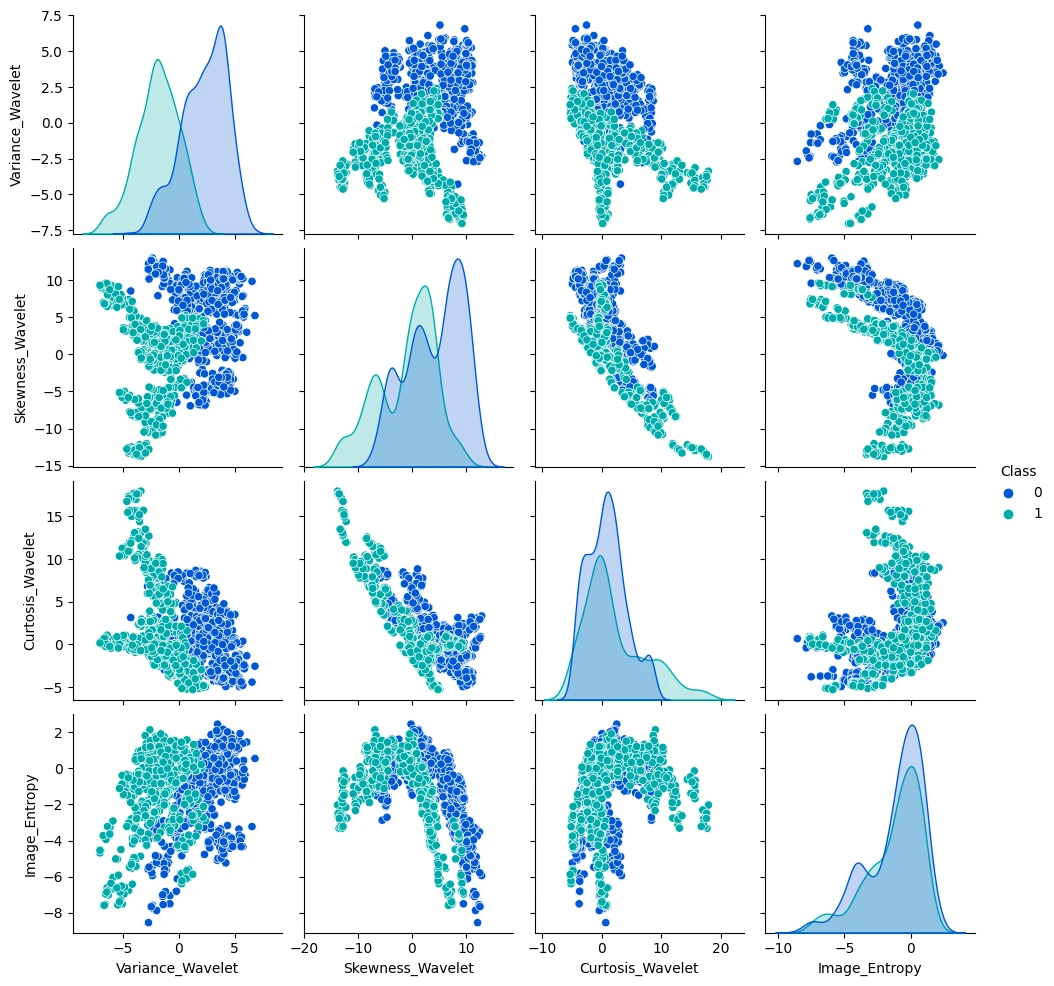
# drop label for training
X_money = money_df.drop('Class', axis=1)
y_money = money_df['Class']
print(X_money.shape, y_money.shape)
X_money_train, X_money_test, y_money_train, y_money_test = train_test_split(
X_money,
y_money,
test_size=0.15,
random_state=42
)
Grid Search for Hyperparameters
rfc_money_base = RandomForestClassifier(oob_score=True)
param_grid = \{
'n_estimators': [64, 96, 128, 160, 192],
'max_features': [2,3,4],
'bootstrap': [True, False]
\}
grid_money = GridSearchCV(rfc_money_base, param_grid)
grid_money.fit(X_money_train, y_money_train)
grid_money.best_params_
# \{'bootstrap': True, 'max_features': 2, 'n_estimators': 96\}
Model Training and Evaluation
rfc_money = RandomForestClassifier(
bootstrap=True,
max_features=2,
n_estimators=96,
oob_score=True
)
rfc_money.fit(X_money_train, y_money_train)
print('Out-of-Bag Score: ', rfc_money.oob_score_.round(4)*100, '%')
# Out-of-Bag Score: 99.14 %
money_pred = rfc_money.predict(X_money_test)
money_report = classification_report(y_money_test, money_pred)
print(money_report)
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| 0 | 0.99 | 1.00 | 1.00 | 111 |
| 1 | 1.00 | 0.99 | 0.99 | 95 |
| accuracy | 1.00 | 206 | ||
| macro avg | 1.00 | 0.99 | 1.00 | 206 |
| weighted avg | 1.00 | 1.00 | 1.00 | 206 |
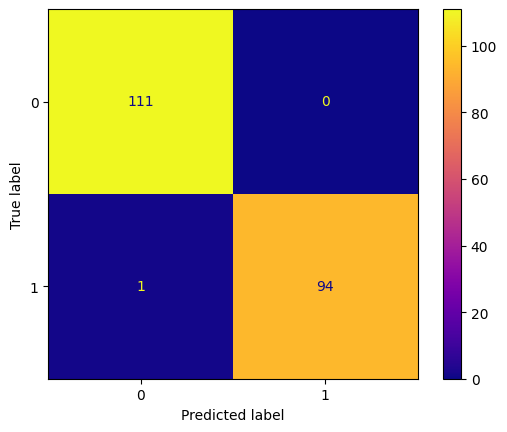
conf_mtx_money = confusion_matrix(y_money_test, money_pred)
conf_mtx_money_plot = ConfusionMatrixDisplay(
confusion_matrix=conf_mtx_money
)
conf_mtx_money_plot.plot(cmap='plasma')
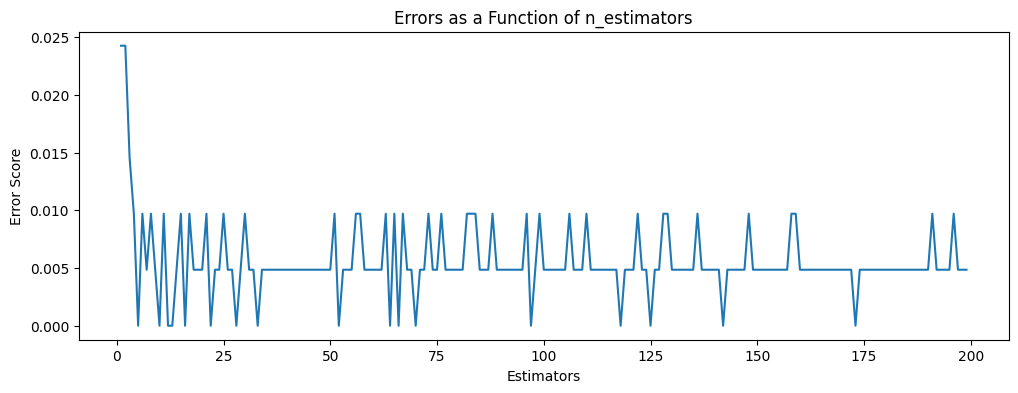
Optimizations
# verify number of estimators found by grid search
errors = []
missclassifications = []
for n in range(1,200):
rfc = RandomForestClassifier(n_estimators=n, max_features=2)
rfc.fit(X_money_train, y_money_train)
preds = rfc.predict(X_money_test)
err = 1 - accuracy_score(y_money_test, preds)
errors.append(err)
n_missed = np.sum(preds != y_money_test)
missclassifications.append(n_missed)
plt.figure(figsize=(12,4))
plt.title('Errors as a Function of n_estimators')
plt.xlabel('Estimators')
plt.ylabel('Error Score')
plt.plot(range(1,200), errors)
# there is no noteable improvement above ~10 estimators
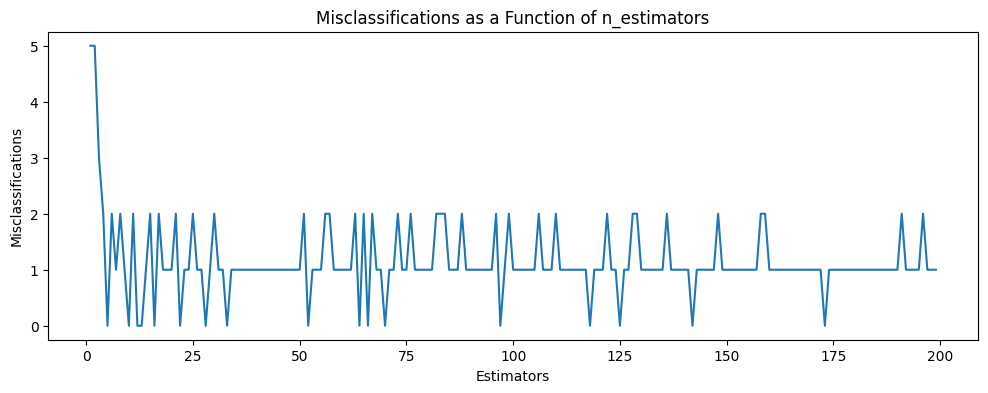
plt.figure(figsize=(12,4))
plt.title('Misclassifications as a Function of n_estimators')
plt.xlabel('Estimators')
plt.ylabel('Misclassifications')
plt.plot(range(1,200), missclassifications)
# and the same for misclassifications
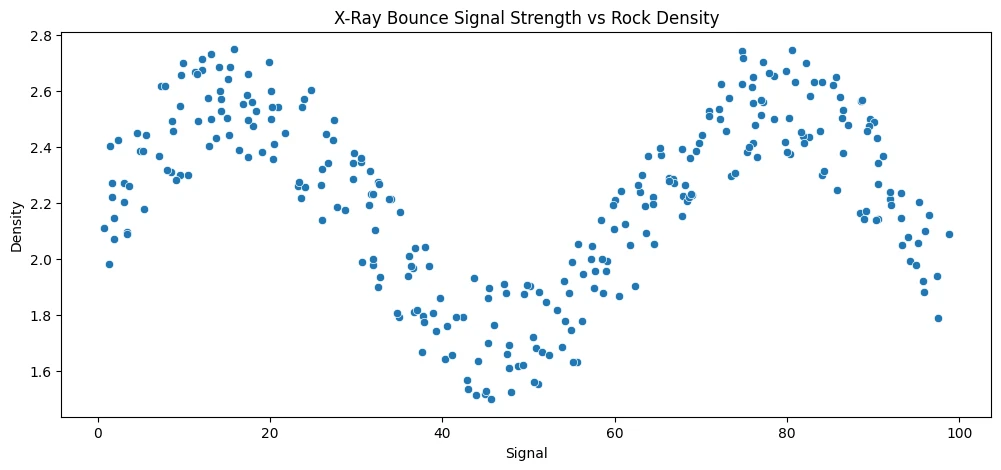
Random Forest Regressor
Comparing different regression models to a random forrest regression model.
# dataset
!wget https://github.com/vineetsingh028/Rock_Density_Prediction/raw/master/rock_density_xray.csv -P datasets
rock_df = pd.read_csv('datasets/rock_density_xray.csv')
rock_df.columns = ['Signal', 'Density']
rock_df.head(5)
| Signal | Density | |
|---|---|---|
| 0 | 72.945124 | 2.456548 |
| 1 | 14.229877 | 2.601719 |
| 2 | 36.597334 | 1.967004 |
| 3 | 9.578899 | 2.300439 |
| 4 | 21.765897 | 2.452374 |
plt.figure(figsize=(12,5))
plt.title('X-Ray Bounce Signal Strength vs Rock Density')
sns.scatterplot(data=rock_df, x='Signal', y='Density')
# the signal vs density plot follows a sine wave - spoiler alert: simpler algorithm
# will fail trying to fit this dataset...
# train-test split
X_rock = rock_df['Signal'].values.reshape(-1,1)
y_rock = rock_df['Density']
X_rock_train, X_rock_test, y_rock_train, y_rock_test = train_test_split(
X_rock,
y_rock,
test_size=0.1,
random_state=42
)
# normalization
scaler = StandardScaler()
X_rock_train_scaled = scaler.fit_transform(X_rock_train)
X_rock_test_scaled = scaler.transform(X_rock_test)
vs Linear Regression
lr_rock = LinearRegression()
lr_rock.fit(X_rock_train_scaled, y_rock_train)
lr_rock_preds = lr_rock.predict(X_rock_test_scaled)
mae = mean_absolute_error(y_rock_test, lr_rock_preds)
rmse = np.sqrt(mean_squared_error(y_rock_test, lr_rock_preds))
mean_abs = y_rock_test.mean()
avg_error = mae * 100 / mean_abs
print('MAE: ', mae.round(2), 'RMSE: ', rmse.round(2), 'Relative Avg. Error: ', avg_error.round(2), '%')
# MAE: 0.24 RMSE: 0.3 Relative Avg. Error: 10.93 %
# visualize predictions
plt.figure(figsize=(12,5))
plt.plot(X_rock_test, lr_rock_preds, c='mediumspringgreen')
sns.scatterplot(data=rock_df, x='Signal', y='Density', c='dodgerblue')
plt.title('Linear Regression Predictions')
plt.show()
# the returned error appears small because the linear regression returns an average
# but it cannot fit a linear line to the contours of the underlying sine wave function
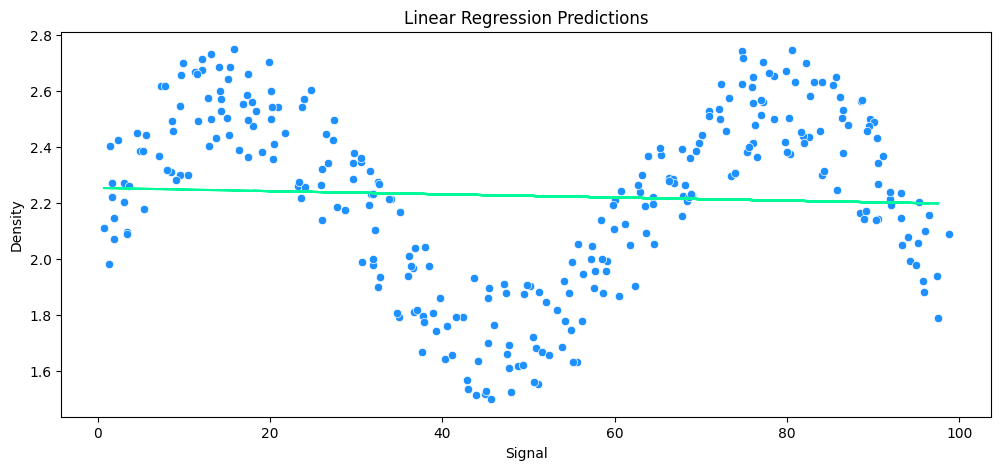
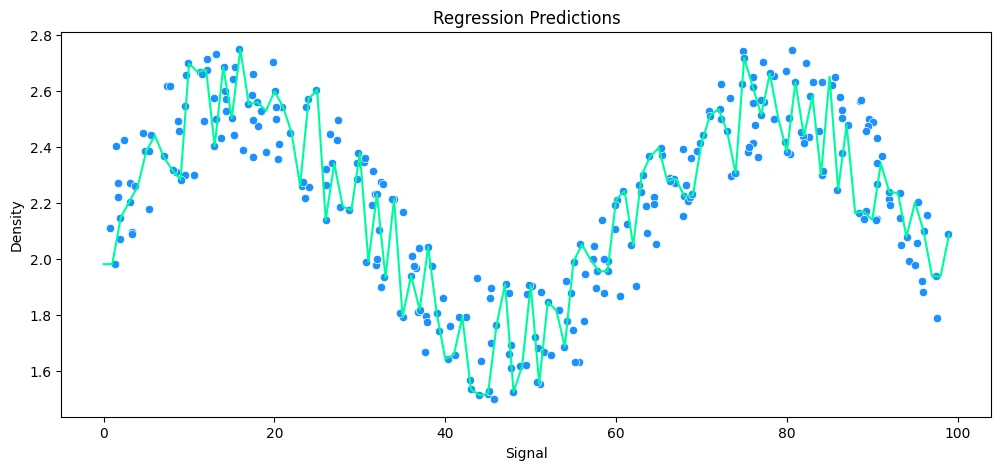
vs Polynomial Regression
# helper function
def run_model(model, X_train, y_train, X_test, y_test, df):
# FIT MODEL
model.fit(X_train, y_train)
# EVALUATE
y_preds = model.predict(X_test)
mae = mean_absolute_error(y_test, y_preds)
rmse = np.sqrt(mean_squared_error(y_test, y_preds))
mean_abs = y_test.mean()
avg_error = mae * 100 / mean_abs
print('MAE: ', mae.round(2), 'RMSE: ', rmse.round(2), 'Relative Avg. Error: ', avg_error.round(2), '%')
# PLOT RESULTS
signal_range = np.arange(0,100)
output = model.predict(signal_range.reshape(-1,1))
plt.figure(figsize=(12,5))
sns.scatterplot(data=df, x='Signal', y='Density', c='dodgerblue')
plt.plot(signal_range,output, c='mediumspringgreen')
plt.title('Regression Predictions')
plt.show()
# test helper on previous linear regression
run_model(
model=lr_rock,
X_train=X_rock_train,
y_train=y_rock_train,
X_test=X_rock_test,
y_test=y_rock_test,
df=rock_df
)
MAE: 0.24 RMSE: 0.3 Relative Avg. Error: 10.93 %
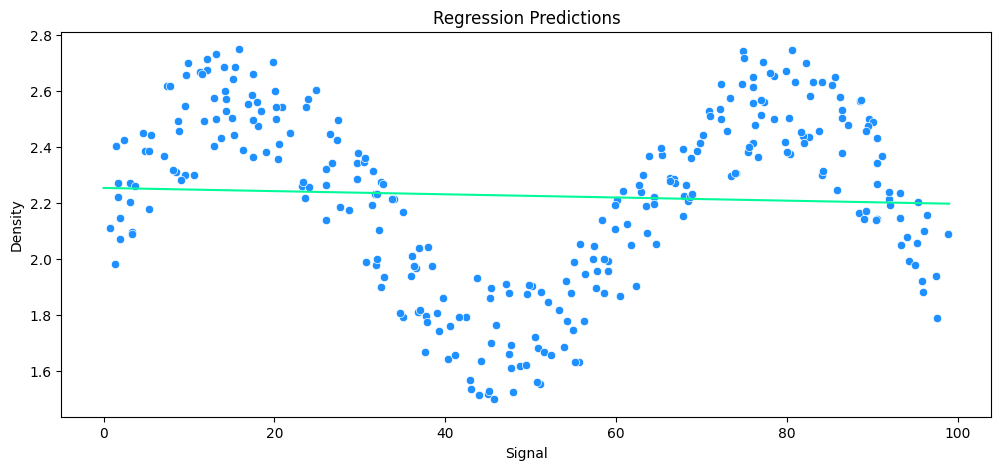
# build polynomial model
pipe_poly = make_pipeline(
PolynomialFeatures(degree=6),
LinearRegression()
)
# run model
run_model(
model=pipe_poly,
X_train=X_rock_train,
y_train=y_rock_train,
X_test=X_rock_test,
y_test=y_rock_test,
df=rock_df
)
# with a HARD LIMIT of 0-100 for the xray signal a 6th degree polinomial is a good fit
MAE: 0.13 RMSE: 0.14 Relative Avg. Error: 5.7 %
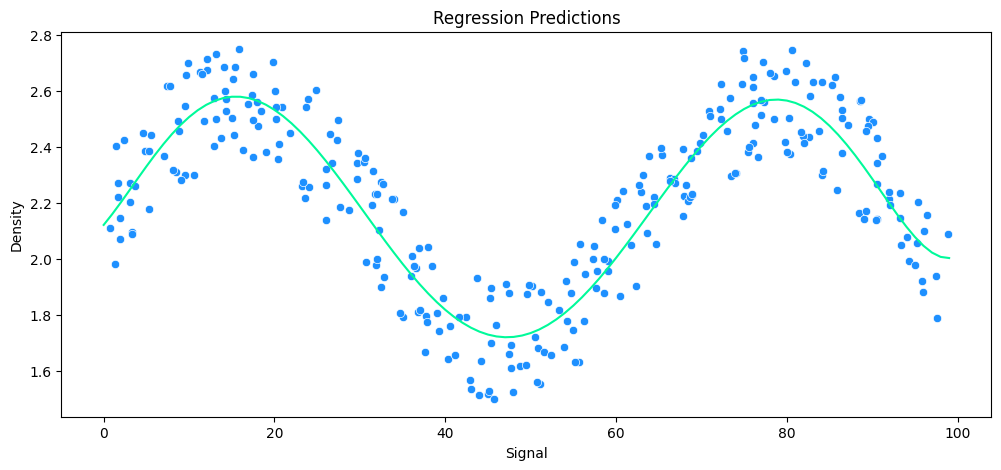
vs KNeighbors Regression
# build polynomial model
k_values=[1,5,10,25]
for k in k_values:
model = KNeighborsRegressor(n_neighbors=k)
print(model)
# run model
run_model(
model,
X_train=X_rock_train,
y_train=y_rock_train,
X_test=X_rock_test,
y_test=y_rock_test,
df=rock_df
)
KNeighborsRegressor(n_neighbors=1)
MAE: 0.12 RMSE: 0.17 Relative Avg. Error: 5.47 %
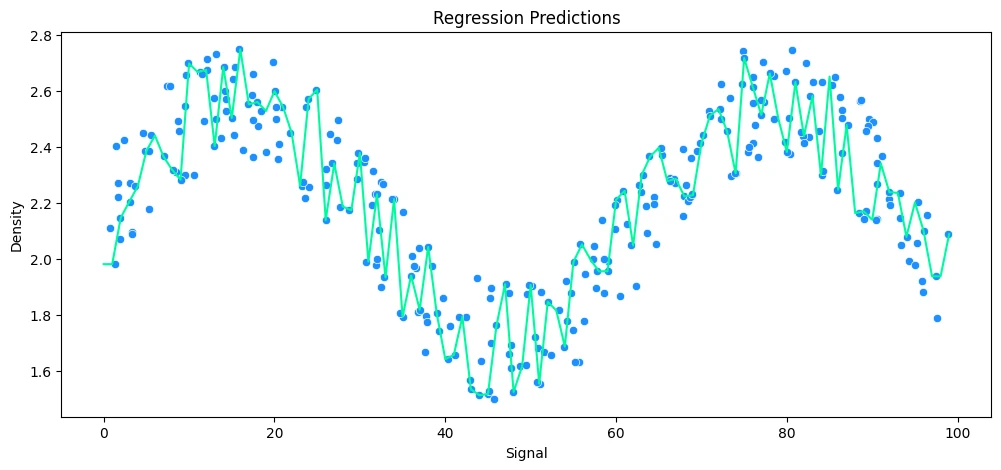
KNeighborsRegressor()
MAE: 0.13 RMSE: 0.15 Relative Avg. Error: 5.9 %
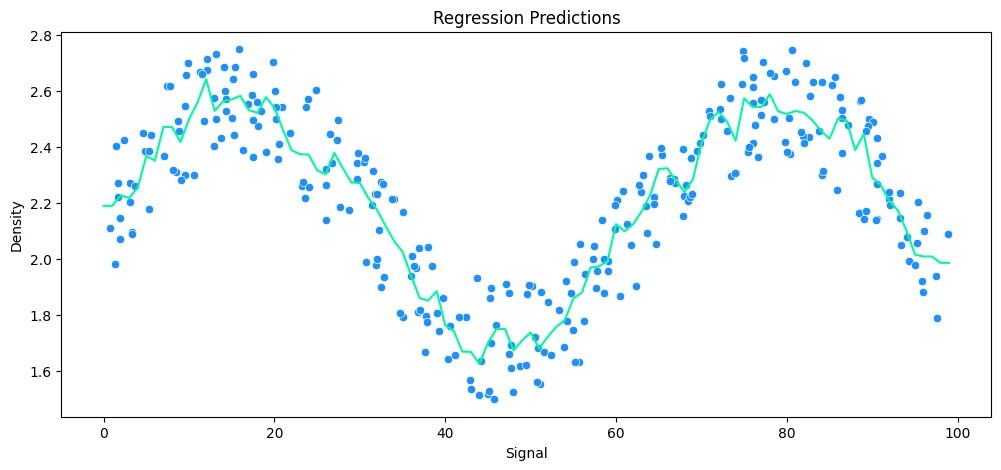
KNeighborsRegressor(n_neighbors=10)
MAE: 0.12 RMSE: 0.14 Relative Avg. Error: 5.44 %
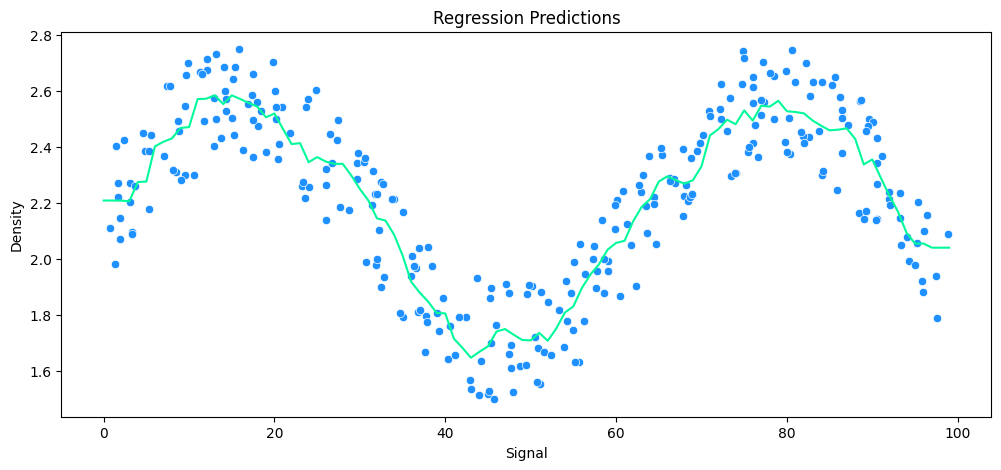
KNeighborsRegressor(n_neighbors=25)
MAE: 0.14 RMSE: 0.16 Relative Avg. Error: 6.18 %
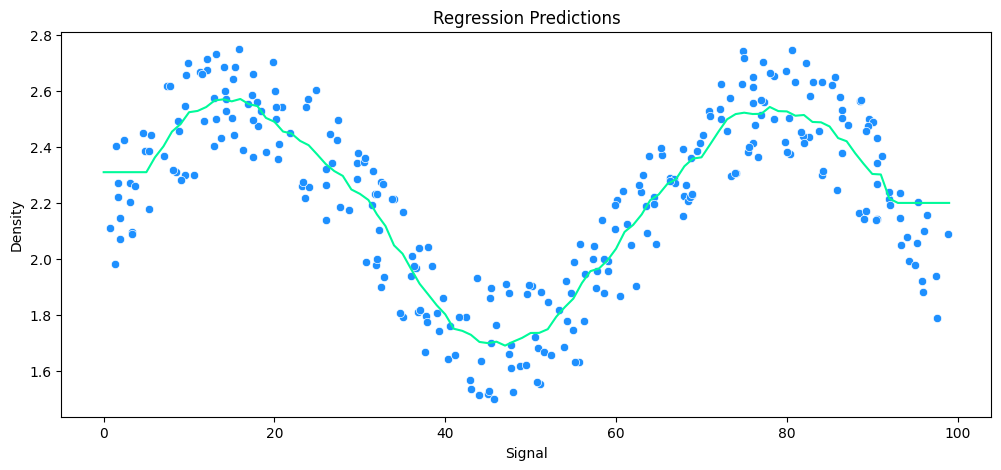
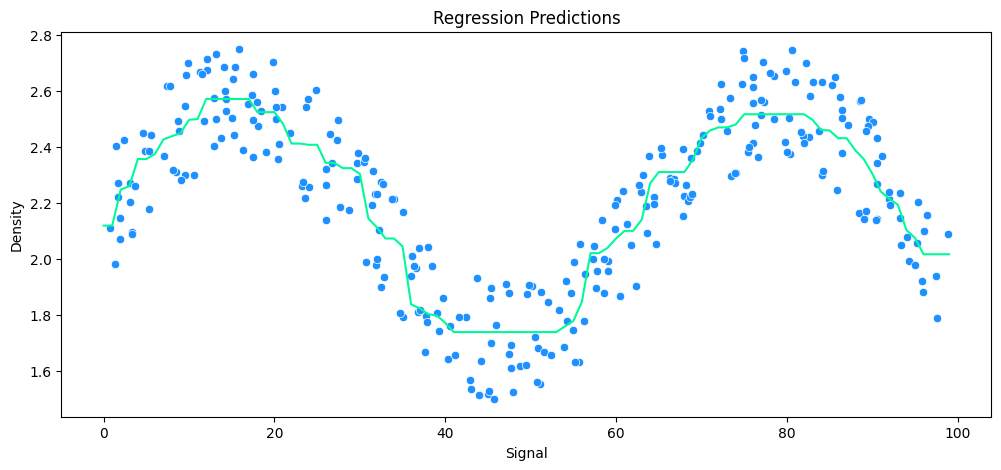
vs Decision Tree Regression
tree_model = DecisionTreeRegressor()
# run model
run_model(
model=tree_model,
X_train=X_rock_train,
y_train=y_rock_train,
X_test=X_rock_test,
y_test=y_rock_test,
df=rock_df
)
MAE: 0.12 RMSE: 0.17 Relative Avg. Error: 5.47 %
vs Support Vector Regression
svr_rock = svm.SVR()
param_grid = \{
'C': [0.01,0.1,1,5,10,100, 1000],
'gamma': ['auto', 'scale']
\}
rock_grid = GridSearchCV(svr_rock, param_grid)
# run model
run_model(
model=rock_grid,
X_train=X_rock_train,
y_train=y_rock_train,
X_test=X_rock_test,
y_test=y_rock_test,
df=rock_df
)
MAE: 0.13 RMSE: 0.14 Relative Avg. Error: 5.75 %
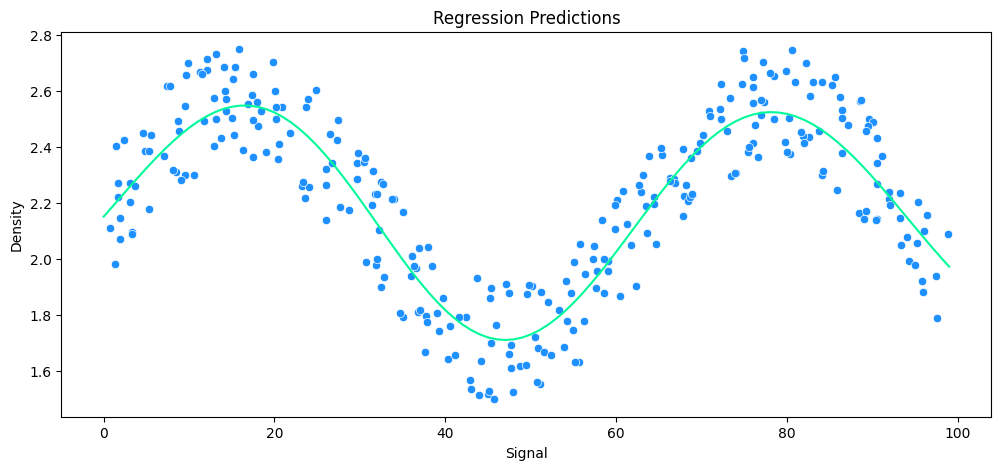
vs Gradient Boosting Regression
gbr_rock = GradientBoostingRegressor()
# run model
run_model(
model=gbr_rock,
X_train=X_rock_train,
y_train=y_rock_train,
X_test=X_rock_test,
y_test=y_rock_test,
df=rock_df
)
MAE: 0.13 RMSE: 0.15 Relative Avg. Error: 5.76 %
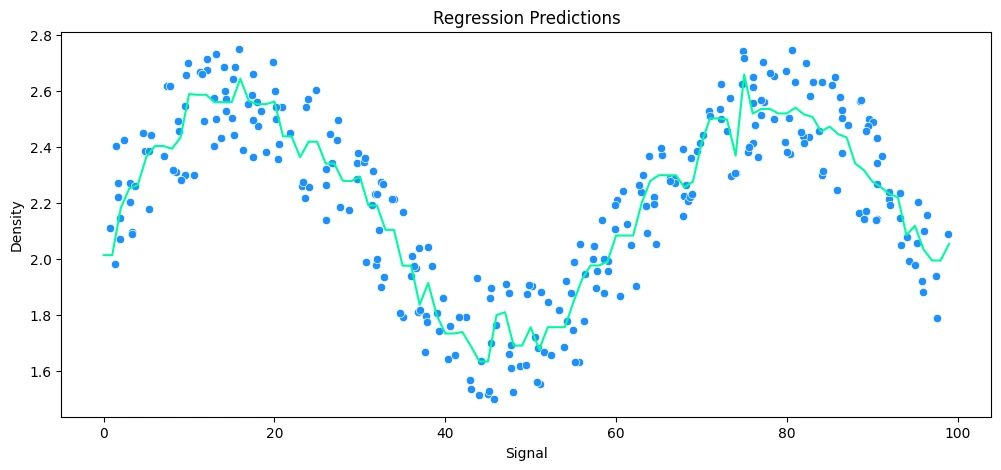
vs Ada Boosting Regression
abr_rock = AdaBoostRegressor()
# run model
run_model(
model=abr_rock,
X_train=X_rock_train,
y_train=y_rock_train,
X_test=X_rock_test,
y_test=y_rock_test,
df=rock_df
)
MAE: 0.13 RMSE: 0.14 Relative Avg. Error: 5.67 %
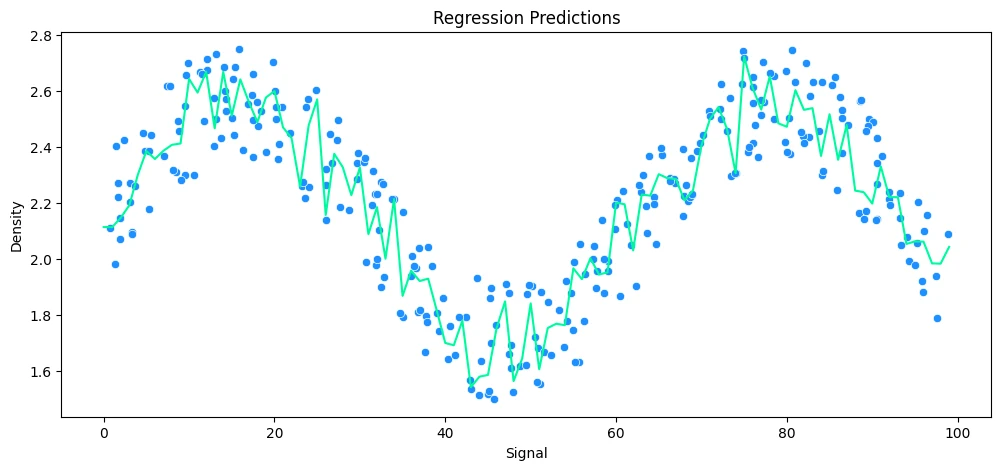
Finally, Random Forrest Regression
rfr_rock = RandomForestRegressor(n_estimators=10)
# run model
run_model(
model=rfr_rock,
X_train=X_rock_train,
y_train=y_rock_train,
X_test=X_rock_test,
y_test=y_rock_test,
df=rock_df
)
MAE: 0.11 RMSE: 0.14 Relative Avg. Error: 5.1 %
Supervised Learning - SVC Model
Support Vector Machines (SVMs) are a set of supervised learning methods used for classification, regression and outliers detection.
- Effective in high dimensional spaces.
- Still effective in cases where number of dimensions is greater than the number of samples.
Dataset
Measurements of geometrical properties of kernels belonging to three different varieties of wheat:
- A: Area,
- P: Perimeter,
- C = 4piA/P^2: Compactness,
- LK: Length of kernel,
- WK: Width of kernel,
- A_Coef: Asymmetry coefficient
- LKG: Length of kernel groove.
!wget https://raw.githubusercontent.com/prasertcbs/basic-dataset/master/Seed_Data.csv -P datasets
wheat_df = pd.read_csv('datasets/Seed_Data.csv')
wheat_df.head(5)
| A | P | C | LK | WK | A_Coef | LKG | target | |
|---|---|---|---|---|---|---|---|---|
| 0 | 15.26 | 14.84 | 0.8710 | 5.763 | 3.312 | 2.221 | 5.220 | 0 |
| 1 | 14.88 | 14.57 | 0.8811 | 5.554 | 3.333 | 1.018 | 4.956 | 0 |
| 2 | 14.29 | 14.09 | 0.9050 | 5.291 | 3.337 | 2.699 | 4.825 | 0 |
| 3 | 13.84 | 13.94 | 0.8955 | 5.324 | 3.379 | 2.259 | 4.805 | 0 |
| 4 | 16.14 | 14.99 | 0.9034 | 5.658 | 3.562 | 1.355 | 5.175 | 0 |
wheat_df.info()
# <class 'pandas.core.frame.DataFrame'>
# RangeIndex: 210 entries, 0 to 209
# Data columns (total 8 columns):
# # Column Non-Null Count Dtype
# --- ------ -------------- -----
# 0 A 210 non-null float64
# 1 P 210 non-null float64
# 2 C 210 non-null float64
# 3 LK 210 non-null float64
# 4 WK 210 non-null float64
# 5 A_Coef 210 non-null float64
# 6 LKG 210 non-null float64
# 7 target 210 non-null int64
# dtypes: float64(7), int64(1)
# memory usage: 13.2 KB
Preprocessing
# remove target feature from training set
X_wheat = wheat_df.drop('target', axis=1)
y_wheat = wheat_df['target']
print(X_wheat.shape, y_wheat.shape)
# (210, 7) (210,)
# train/test split
X_train_wheat, X_test_wheat, y_train_wheat, y_test_wheat = train_test_split(
X_wheat,
y_wheat,
test_size=0.2,
random_state=42
)
# normalization
sc_wheat = StandardScaler()
X_train_wheat=sc_wheat.fit_transform(X_train_wheat)
X_test_wheat=sc_wheat.fit_transform(X_test_wheat)
Model Training
# SVM classifier fitting
clf_wheat = svm.SVC()
clf_wheat.fit(X_train_wheat, y_train_wheat)
Model Evaluation
# Predictions
y_wheat_pred = clf_wheat.predict(X_test_wheat)
print(
'Accuracy Score: ',
accuracy_score(y_test_wheat, y_wheat_pred, normalize=True).round(4)*100, '%'
)
# Accuracy Score: 90.48 %
report_wheat = classification_report(
y_test_wheat, y_wheat_pred
)
print(report_wheat)
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| 0 | 0.82 | 0.82 | 0.82 | 11 |
| 1 | 1.00 | 0.93 | 0.96 | 14 |
| 2 | 0.89 | 0.94 | 0.91 | 17 |
| accuracy | 0.90 | 42 | ||
| macro avg | 0.90 | 0.90 | 0.90 | 42 |
| weighted avg | 0.91 | 0.90 | 0.91 | 42 |
conf_mtx_wheat = confusion_matrix(y_test_wheat, y_wheat_pred)
conf_mtx_wheat
# array([[ 9, 0, 2],
# [ 1, 13, 0],
# [ 1, 0, 16]])
conf_mtx_wheat_plot = ConfusionMatrixDisplay(
confusion_matrix=conf_mtx_wheat
)
conf_mtx_wheat_plot.plot()
plt.show()
Margin Plots for Support Vector Classifier
# get dataset
!wget https://github.com/alpeshraj/mouse_viral_study/raw/main/mouse_viral_study.csv -P datasets
mice_df = pd.read_csv('datasets/mouse_viral_study.csv')
mice_df.head(5)
| Med_1_mL | Med_2_mL | Virus Present | |
|---|---|---|---|
| 0 | 6.508231 | 8.582531 | 0 |
| 1 | 4.126116 | 3.073459 | 1 |
| 2 | 6.427870 | 6.369758 | 0 |
| 3 | 3.672953 | 4.905215 | 1 |
| 4 | 1.580321 | 2.440562 | 1 |
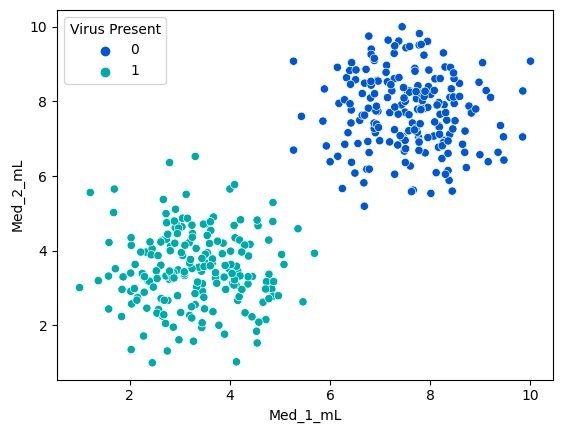
sns.scatterplot(data=mice_df, x='Med_1_mL',y='Med_2_mL',hue='Virus Present', palette='winter')
# visualizing a hyperplane to separate the two features
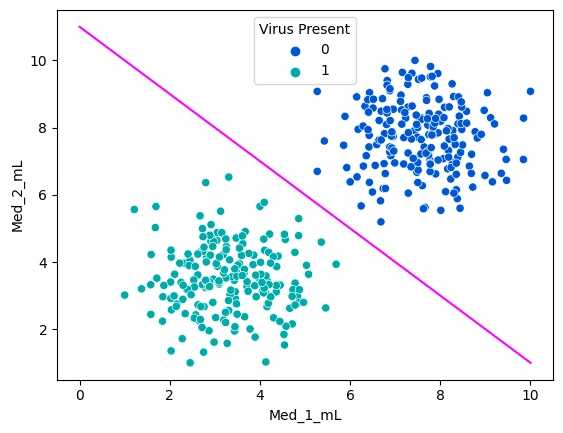
sns.scatterplot(data=mice_df, x='Med_1_mL',y='Med_2_mL',hue='Virus Present', palette='winter')
x = np.linspace(0,10,100)
m = -1
b = 11
y = m*x + b
plt.plot(x,y,c='fuchsia')
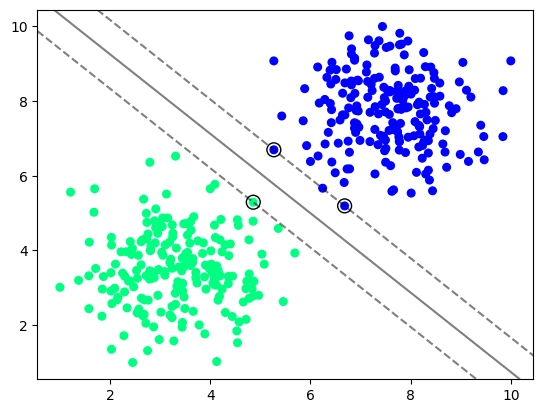
SVC with a Linear Kernel
# using a support vector classifier to calculate maximize the margin between both classes
y_vir = mice_df['Virus Present']
X_vir = mice_df.drop('Virus Present',axis=1)
# kernel : \{'linear', 'poly', 'rbf', 'sigmoid', 'precomputed'\}
# the smaller the C value the more feature vectors will be inside the margin
model_vir = svm.SVC(kernel='linear', C=1000)
model_vir.fit(X_vir, y_vir)
# import helper function
from helper.svm_margin_plot import plot_svm_boundary
plot_svm_boundary(model_vir, X_vir, y_vir)
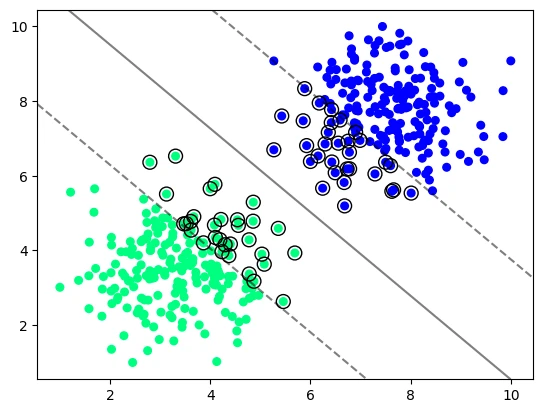
# the smaller the C value the more feature vectors will be inside the margin
model_vir_low_reg = svm.SVC(kernel='linear', C=0.005)
model_vir_low_reg.fit(X_vir, y_vir)
plot_svm_boundary(model_vir_low_reg, X_vir, y_vir)
SVC with a Radial Basis Function Kernel
model_vir_rbf = svm.SVC(kernel='rbf', C=1)
model_vir_rbf.fit(X_vir, y_vir)
plot_svm_boundary(model_vir_rbf, X_vir, y_vir)
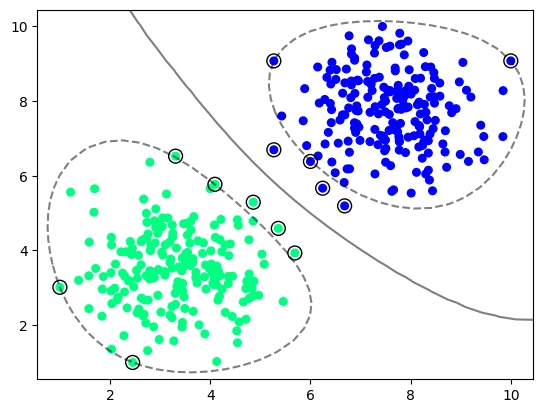
# # gamma : \{'scale', 'auto'\} or float, default='scale'
# - if ``gamma='scale'`` (default) is passed then it uses 1 / (n_features * X.var()) as value of gamma,
# - if 'auto', uses 1 / n_features
# - if float, must be non-negative.
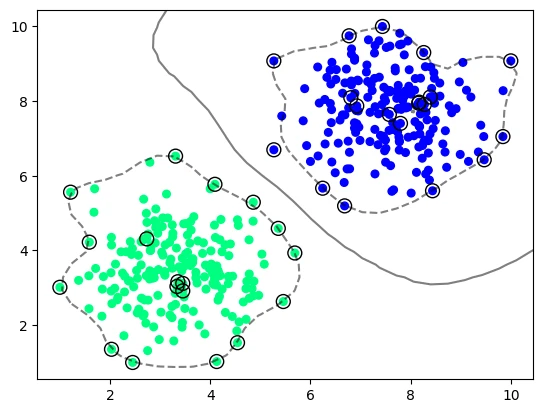
model_vir_rbf_auto_gamma = svm.SVC(kernel='rbf', C=1, gamma='auto')
model_vir_rbf_auto_gamma.fit(X_vir, y_vir)
plot_svm_boundary(model_vir_rbf_auto_gamma, X_vir, y_vir)
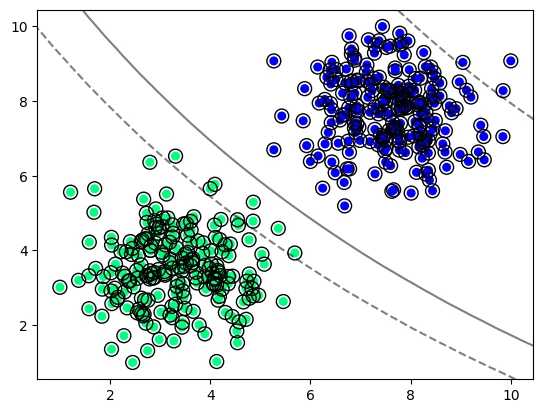
SVC with a Sigmoid Kernel
model_vir_sigmoid = svm.SVC(kernel='sigmoid', gamma='scale')
model_vir_sigmoid.fit(X_vir, y_vir)
plot_svm_boundary(model_vir_sigmoid, X_vir, y_vir)
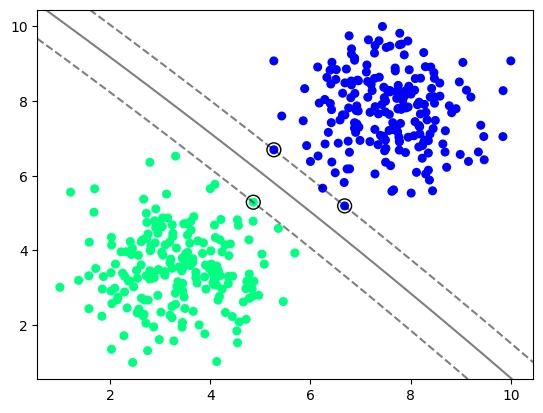
SVC with a Polynomial Kernel
model_vir_poly = svm.SVC(kernel='poly', C=1, degree=2)
model_vir_poly.fit(X_vir, y_vir)
plot_svm_boundary(model_vir_poly, X_vir, y_vir)
Grid Search for Support Vector Classifier
svm_base_model = svm.SVC()
param_grid = \{
'C':[0.01, 0.1, 1],
'kernel': ['linear', 'rbf']
\}
grid = GridSearchCV(svm_base_model, param_grid)
grid.fit(X_vir, y_vir)
grid.best_params_
# \{'C': 0.01, 'kernel': 'linear'\}
Support Vector Regression
# dataset
!wget https://github.com/fsdhakan/ML/raw/main/cement_slump.csv -P datasets
cement_df = pd.read_csv('datasets/cement_slump.csv')
cement_df.head(5)
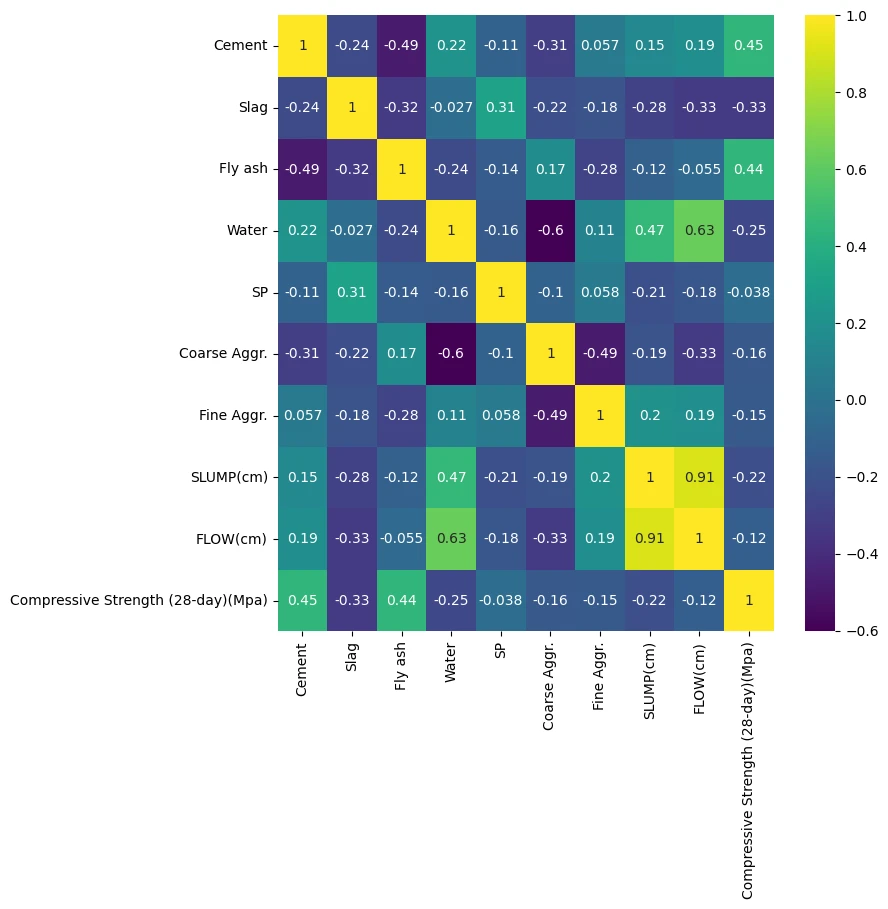
| Cement | Slag | Fly ash | Water | SP | Coarse Aggr. | Fine Aggr. | SLUMP(cm) | FLOW(cm) | Compressive Strength (28-day)(Mpa) | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 273.0 | 82.0 | 105.0 | 210.0 | 9.0 | 904.0 | 680.0 | 23.0 | 62.0 | 34.99 |
| 1 | 163.0 | 149.0 | 191.0 | 180.0 | 12.0 | 843.0 | 746.0 | 0.0 | 20.0 | 41.14 |
| 2 | 162.0 | 148.0 | 191.0 | 179.0 | 16.0 | 840.0 | 743.0 | 1.0 | 20.0 | 41.81 |
| 3 | 162.0 | 148.0 | 190.0 | 179.0 | 19.0 | 838.0 | 741.0 | 3.0 | 21.5 | 42.08 |
| 4 | 154.0 | 112.0 | 144.0 | 220.0 | 10.0 | 923.0 | 658.0 | 20.0 | 64.0 | 26.82 |
plt.figure(figsize=(8,8))
sns.heatmap(cement_df.corr(), annot=True, cmap='viridis')
# drop labels
X_cement = cement_df.drop('Compressive Strength (28-day)(Mpa)', axis=1)
y_cement = cement_df['Compressive Strength (28-day)(Mpa)']
# train/test split
X_train_cement, X_test_cement, y_train_cement, y_test_cement = train_test_split(
X_cement,
y_cement,
test_size=0.3,
random_state=42
)
# normalize
scaler = StandardScaler()
X_train_cement_scaled = scaler.fit_transform(X_train_cement)
X_test_cement_scaled = scaler.transform(X_test_cement)
Base Model Run
base_model_cement = svm.SVR()
base_model_cement.fit(X_train_cement_scaled, y_train_cement)
base_model_predictions = base_model_cement.predict(X_test_cement_scaled)
mae = mean_absolute_error(y_test_cement, base_model_predictions)
rmse = mean_squared_error(y_test_cement, base_model_predictions)
mean_abs = y_test_cement.mean()
avg_error = mae * 100 / mean_abs
print('MAE: ', mae.round(2), 'RMSE: ', rmse.round(2), 'Relative Avg. Error: ', avg_error.round(2), '%')
| MAE | RMSE | Relative Avg. Error |
|---|---|---|
| 4.68 | 36.95 | 12.75 % |
Grid Search for better Hyperparameter
param_grid = \{
'C': [0.001,0.01,0.1,0.5,1],
'kernel': ['linear', 'rbf', 'poly'],
'gamma': ['scale', 'auto'],
'degree': [2,3,4],
'epsilon': [0,0.01,0.1,0.5,1,2]
\}
cement_grid = GridSearchCV(base_model_cement, param_grid)
cement_grid.fit(X_train_cement_scaled, y_train_cement)
cement_grid.best_params_
# \{'C': 1, 'degree': 2, 'epsilon': 2, 'gamma': 'scale', 'kernel': 'linear'\}
cement_grid_predictions = cement_grid.predict(X_test_cement_scaled)
mae_grid = mean_absolute_error(y_test_cement, cement_grid_predictions)
rmse_grid = mean_squared_error(y_test_cement, cement_grid_predictions)
mean_abs = y_test_cement.mean()
avg_error_grid = mae_grid * 100 / mean_abs
print('MAE: ', mae_grid.round(2), 'RMSE: ', rmse_grid.round(2), 'Relative Avg. Error: ', avg_error_grid.round(2), '%')
| MAE | RMSE | Relative Avg. Error |
|---|---|---|
| 1.85 | 5.2 | 5.05 % |
Example Task - Wine Fraud
Data Exploration
# dataset
!wget https://github.com/CAPGAGA/Fraud-in-Wine/raw/main/wine_fraud.csv -P datasets
wine_df = pd.read_csv('datasets/wine_fraud.csv')
wine_df.head(5)
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | quality | type | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 7.4 | 0.70 | 0.00 | 1.9 | 0.076 | 11.0 | 34.0 | 0.9978 | 3.51 | 0.56 | 9.4 | Legit | red |
| 1 | 7.8 | 0.88 | 0.00 | 2.6 | 0.098 | 25.0 | 67.0 | 0.9968 | 3.20 | 0.68 | 9.8 | Legit | red |
| 2 | 7.8 | 0.76 | 0.04 | 2.3 | 0.092 | 15.0 | 54.0 | 0.9970 | 3.26 | 0.65 | 9.8 | Legit | red |
| 3 | 11.2 | 0.28 | 0.56 | 1.9 | 0.075 | 17.0 | 60.0 | 0.9980 | 3.16 | 0.58 | 9.8 | Legit | red |
| 4 | 7.4 | 0.70 | 0.00 | 1.9 | 0.076 | 11.0 | 34.0 | 0.9978 | 3.51 | 0.56 | 9.4 | Legit | red |
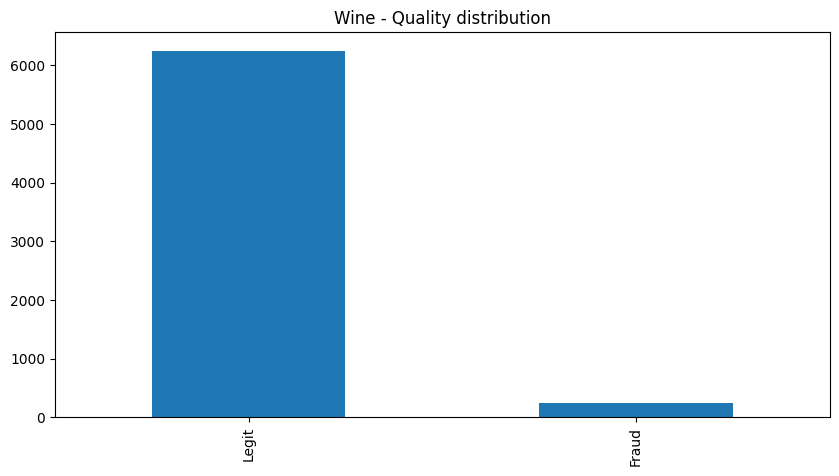
wine_df.value_counts('quality')
| quality | |
|---|---|
| Legit | 6251 |
| Fraud | 246 |
| dtype: int64 |
wine_df['quality'].value_counts().plot(
kind='bar',
figsize=(10,5),
title='Wine - Quality distribution')
plt.figure(figsize=(10, 5))
plt.title('Wine - Quality distribution by Type')
sns.countplot(
data=wine_df,
x='quality',
hue='type',
palette='winter'
)
plt.savefig('assets/Scikit_Learn_22.webp', bbox_inches='tight')
wine_df_white = wine_df[wine_df['type'] == 'white']
wine_df_red = wine_df[wine_df['type'] == 'red']
# fraud percentage by wine type
legit_white_wines = wine_df_white.value_counts('quality')[0]
fraud_white_wines = wine_df_white.value_counts('quality')[1]
white_fraud_percentage = fraud_white_wines * 100 / (legit_white_wines + fraud_white_wines)
legit_red_wines = wine_df_red.value_counts('quality')[0]
fraud_red_wines = wine_df_red.value_counts('quality')[1]
red_fraud_percentage = fraud_red_wines * 100 / (legit_red_wines + fraud_red_wines)
print(
'Fraud Percentage: \nWhite Wines: ',
white_fraud_percentage.round(2),
'% \nRed Wines: ',
red_fraud_percentage.round(2),
'%'
)
| Fraud Percentage: | |
|---|---|
| White Wines: | 3.74 % |
| Red Wines: | 3.94 % |
# make features numeric
feature_map = \{
'Legit': 0,
'Fraud': 1,
'red': 0,
'white': 1
\}
wine_df['quality_enc'] = wine_df['quality'].map(feature_map)
wine_df['type_enc'] = wine_df['type'].map(feature_map)
wine_df[['quality', 'quality_enc', 'type', 'type_enc']]
| quality | quality_enc | type | type_enc | |
|---|---|---|---|---|
| 0 | Legit | 0 | red | 0 |
| 1 | Legit | 0 | red | 0 |
| 2 | Legit | 0 | red | 0 |
| 3 | Legit | 0 | red | 0 |
| 4 | Legit | 0 | red | 0 |
| ... | ||||
| 6492 | Legit | 0 | white | 1 |
| 6493 | Legit | 0 | white | 1 |
| 6494 | Legit | 0 | white | 1 |
| 6495 | Legit | 0 | white | 1 |
| 6496 | Legit | 0 | white | 1 |
| 6497 rows × 4 columns |
# find correlations
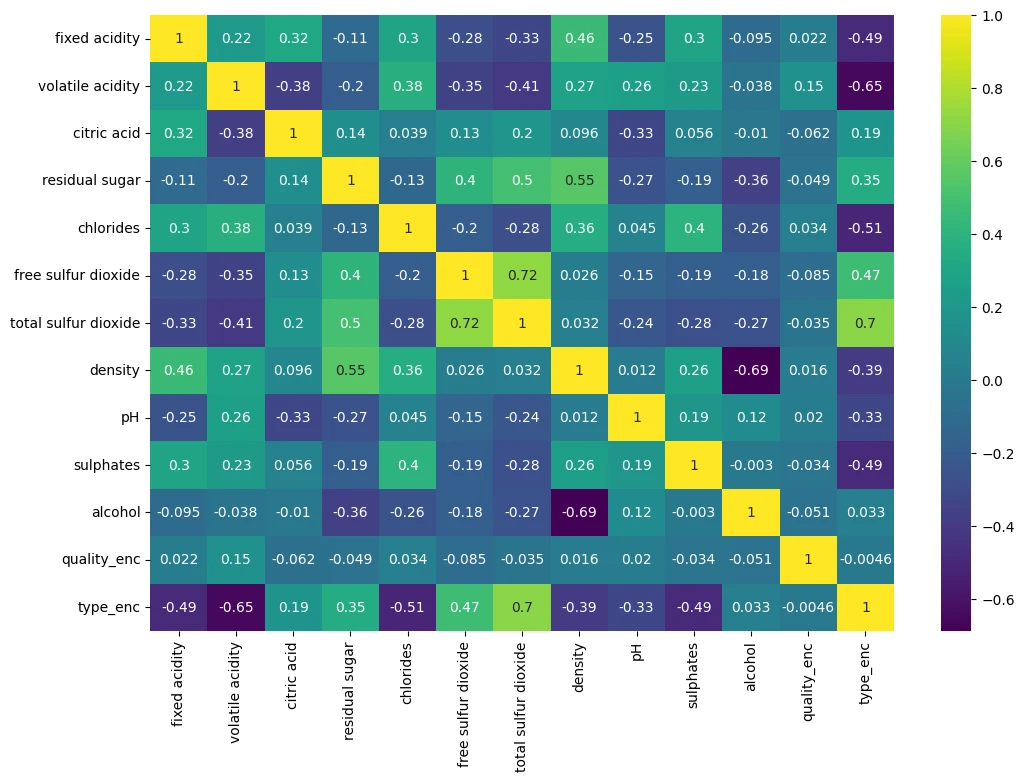
wine_df.corr(numeric_only=True)
| fixed acidity | volatile acidity | citric acid | residual sugar | chlorides | free sulfur dioxide | total sulfur dioxide | density | pH | sulphates | alcohol | quality_enc | type_enc | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| fixed acidity | 1.000000 | 0.219008 | 0.324436 | -0.111981 | 0.298195 | -0.282735 | -0.329054 | 0.458910 | -0.252700 | 0.299568 | -0.095452 | 0.021794 | -0.486740 |
| volatile acidity | 0.219008 | 1.000000 | -0.377981 | -0.196011 | 0.377124 | -0.352557 | -0.414476 | 0.271296 | 0.261454 | 0.225984 | -0.037640 | 0.151228 | -0.653036 |
| citric acid | 0.324436 | -0.377981 | 1.000000 | 0.142451 | 0.038998 | 0.133126 | 0.195242 | 0.096154 | -0.329808 | 0.056197 | -0.010493 | -0.061789 | 0.187397 |
| residual sugar | -0.111981 | -0.196011 | 0.142451 | 1.000000 | -0.128940 | 0.402871 | 0.495482 | 0.552517 | -0.267320 | -0.185927 | -0.359415 | -0.048756 | 0.348821 |
| chlorides | 0.298195 | 0.377124 | 0.038998 | -0.128940 | 1.000000 | -0.195045 | -0.279630 | 0.362615 | 0.044708 | 0.395593 | -0.256916 | 0.034499 | -0.512678 |
| free sulfur dioxide | -0.282735 | -0.352557 | 0.133126 | 0.402871 | -0.195045 | 1.000000 | 0.720934 | 0.025717 | -0.145854 | -0.188457 | -0.179838 | -0.085204 | 0.471644 |
| total sulfur dioxide | -0.329054 | -0.414476 | 0.195242 | 0.495482 | -0.279630 | 0.720934 | 1.000000 | 0.032395 | -0.238413 | -0.275727 | -0.265740 | -0.035252 | 0.700357 |
| density | 0.458910 | 0.271296 | 0.096154 | 0.552517 | 0.362615 | 0.025717 | 0.032395 | 1.000000 | 0.011686 | 0.259478 | -0.686745 | 0.016351 | -0.390645 |
| pH | -0.252700 | 0.261454 | -0.329808 | -0.267320 | 0.044708 | -0.145854 | -0.238413 | 0.011686 | 1.000000 | 0.192123 | 0.121248 | 0.020107 | -0.329129 |
| sulphates | 0.299568 | 0.225984 | 0.056197 | -0.185927 | 0.395593 | -0.188457 | -0.275727 | 0.259478 | 0.192123 | 1.000000 | -0.003029 | -0.034046 | -0.487218 |
| alcohol | -0.095452 | -0.037640 | -0.010493 | -0.359415 | -0.256916 | -0.179838 | -0.265740 | -0.686745 | 0.121248 | -0.003029 | 1.000000 | -0.051141 | 0.032970 |
| quality_enc | 0.021794 | 0.151228 | -0.061789 | -0.048756 | 0.034499 | -0.085204 | -0.035252 | 0.016351 | 0.020107 | -0.034046 | -0.051141 | 1.000000 | -0.004598 |
| type_enc | -0.486740 | -0.653036 | 0.187397 | 0.348821 | -0.512678 | 0.471644 | 0.700357 | -0.390645 | -0.329129 | -0.487218 | 0.032970 | -0.004598 | 1.000000 |
plt.figure(figsize=(12,8))
sns.heatmap(wine_df.corr(numeric_only=True), annot=True, cmap='viridis')
# how does the quality correlate to measurements
wine_df.corr(numeric_only=True)['quality_enc']
| Quality Correlstion | |
|---|---|
| fixed acidity | 0.021794 |
| volatile acidity | 0.151228 |
| citric acid | -0.061789 |
| residual sugar | -0.048756 |
| chlorides | 0.034499 |
| free sulfur dioxide | -0.085204 |
| total sulfur dioxide | -0.035252 |
| density | 0.016351 |
| pH | 0.020107 |
| sulphates | -0.034046 |
| alcohol | -0.051141 |
| quality_enc | 1.000000 |
| type_enc | -0.004598 |
| Name: quality_enc, dtype: float64 |
wine_df.corr(numeric_only=True)['quality_enc'][:-2].sort_values().plot(
figsize=(12,5),
kind='bar',
title='Correlation of Measurements to Quality'
)
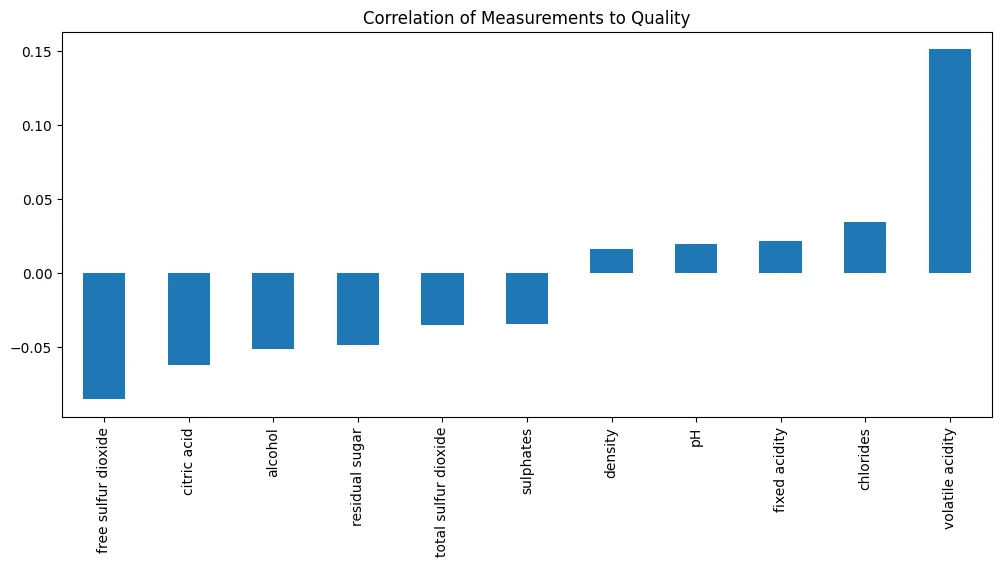
Regression Model
# separate target + remove string values
X_wine = wine_df.drop(['quality_enc', 'quality', 'type'], axis=1)
y_wine = wine_df['quality']
print(X_wine.shape, y_wine.shape)
# train-test split
X_wine_train, X_wine_test, y_wine_train, y_wine_test = train_test_split(
X_wine,
y_wine,
test_size=0.1,
random_state=42
)
# normalization
scaler = StandardScaler()
X_wine_train_scaled = scaler.fit_transform(X_wine_train)
X_wine_test_scaled = scaler.transform(X_wine_test)
# create the SVC model using class_weight to balance out the
# dataset that heavily leaning towards non-frauds
svc_wine_base = svm.SVC(
kernel='rbf',
class_weight='balanced'
)
# grid search
param_grid = \{
'C': [0.5, 1, 1.5, 2, 2.5],
'gamma' : ['scale', 'auto']
\}
wine_grid = GridSearchCV(svc_wine_base, param_grid)
wine_grid.fit(X_wine_train_scaled, y_wine_train)
print('Best Params: ', wine_grid.best_params_)
# Best Params: \{'C': 2.5, 'gamma': 'auto'\}
y_wine_pred = wine_grid.predict(X_wine_test_scaled)
print(
'Accuracy Score: ',
accuracy_score(y_wine_test, y_wine_pred, normalize=True).round(4)*100, '%'
)
# Accuracy Score: 84.77 %
report_wine = classification_report(
y_wine_test, y_wine_pred
)
print(report_wine)
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| Fraud | 0.16 | 0.68 | 0.26 | 25 |
| Legit | 0.99 | 0.85 | 0.92 | 625 |
| accuracy | 0.85 | 650 | ||
| macro avg | 0.57 | 0.77 | 0.59 | 650 |
| weighted avg | 0.95 | 0.85 | 0.89 | 650 |
conf_mtx_wine = confusion_matrix(y_wine_test, y_wine_pred)
conf_mtx_wine
# array([[ 17, 8],
# [ 91, 534]])
conf_mtx_wine_plot = ConfusionMatrixDisplay(
confusion_matrix=conf_mtx_wine
)
conf_mtx_wine_plot.plot(cmap='plasma')
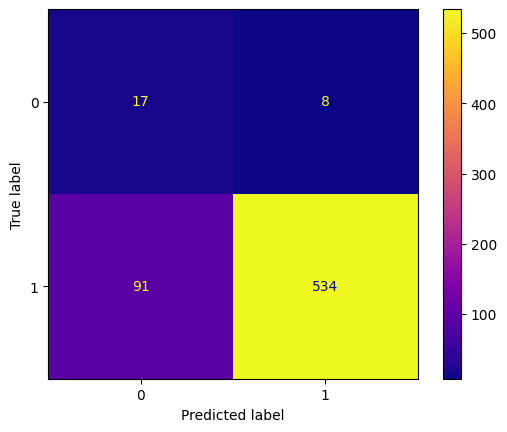
# expand grid search
param_grid = \{
'C': [1000, 1050, 1100, 1050, 1200],
'gamma' : ['scale', 'auto']
\}
wine_grid = GridSearchCV(svc_wine_base, param_grid)
wine_grid.fit(X_wine_train_scaled, y_wine_train)
print('Best Params: ', wine_grid.best_params_)
# Best Params: \{'C': 1100, 'gamma': 'scale'\}
y_wine_pred = wine_grid.predict(X_wine_test_scaled)
print('Accuracy Score: ',accuracy_score(y_wine_test, y_wine_pred, normalize=True).round(4)*100, '%')
# Accuracy Score: 94.31 %
report_wine = classification_report(y_wine_test, y_wine_pred)
print(report_wine)
conf_mtx_wine = confusion_matrix(y_wine_test, y_wine_pred)
conf_mtx_wine_plot = ConfusionMatrixDisplay(
confusion_matrix=conf_mtx_wine
)
conf_mtx_wine_plot.plot(cmap='plasma')
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| Fraud | 0.29 | 0.32 | 0.30 | 25 |
| Legit | 0.97 | 0.97 | 0.97 | 625 |
| accuracy | 0.85 | 650 | ||
| macro avg | 0.63 | 0.64 | 0.64 | 650 |
| weighted avg | 0.95 | 0.94 | 0.94 | 650 |
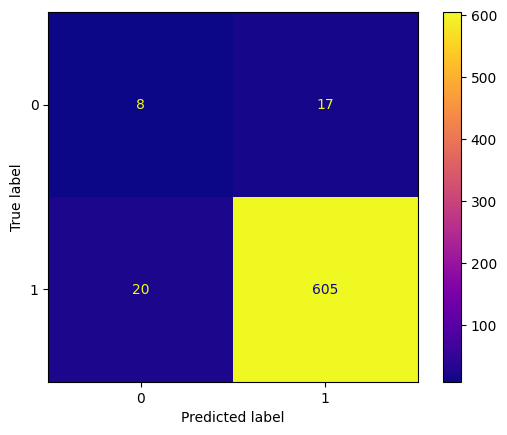
Supervised Learning - Boosting Methods
# dataset - label mushrooms as poisonous or eatable
!wget https://github.com/semnan-university-ai/Mushroom/raw/main/Mushroom.csv -P datasets
Dataset Exploration
shroom_df = pd.read_csv('datasets/mushrooms.csv')
shroom_df.head(5).transpose()
- cap-shape: bell =
b, conical =c, convex =x, flat =f, knobbed =k, sunken =s - cap-surface: fibrous =
f, grooves =g, scaly =y, smooth =s - cap-color: brown =
n, buff =b, cinnamon =c, gray =g, green =r, pink =p, purple =u, red =e, white =w, yellow =y - bruises?: bruises =
t, no =f - odor: almond =
a, anise =l, creosote =c, fishy =y, foul =f, musty =m, none =n, pungent =p, spicy =s - gill-attachment: attached =
a, descending =d, free =f, notched =n - gill-spacing: close =
c, crowded =w, distant =d - gill-size: broad =
b, narrow =n - gill-color: black =
k, brown =n, buff =b, chocolate =h, gray =g, green =r, orange =o, pink =p, purple =u, red =e, white =w, yellow =y - stalk-shape: enlarging =
e, tapering =t - stalk-root: bulbous =
b, club =c, cup =u, equal =e, rhizomorphs =z, rooted =r, missing =? - stalk-surface-above-ring: fibrous =
f, scaly =y, silky =k, smooth =s - stalk-surface-below-ring: fibrous =
f, scaly =y, silky =k, smooth =s - stalk-color-above-ring: brown =
n, buff =b, cinnamon =c, gray =g, orange =o, pink =p, red =e, white =w, yellow =y - stalk-color-below-ring: brown =
n, buff =b, cinnamon =c, gray =g, orange =o, pink =p, red =e, white =w, yellow =y - veil-type: partial =
p, universal =u - veil-color: brown =
n, orange =o, white =w, yellow =y - ring-number: none =
n, one =o, two =t - ring-type: cobwebby =
c, evanescent =e, flaring =f, large =l, none =n, pendant =p, sheathing =s, zone =z - spore-print-color: black =
k, brown =n, buff =b, chocolate =h, green =r, orange =o, purple =u, white =w, yellow =y - population: abundant =
a, clustered =c, numerous =n, scattered =s, several =v, solitary = `y - habitat: grasses =
g, leaves =l, meadows =m, paths =p, urban =u, waste =w, woods =d
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| class | p | e | e | p | e |
| cap-shape | x | x | b | x | x |
| cap-surface | s | s | s | y | s |
| cap-color | n | y | w | w | g |
| bruises | t | t | t | t | f |
| odor | p | a | l | p | n |
| gill-attachment | f | f | f | f | f |
| gill-spacing | c | c | c | c | w |
| gill-size | n | b | b | n | b |
| gill-color | k | k | n | n | k |
| stalk-shape | e | e | e | e | t |
| stalk-root | e | c | c | e | e |
| stalk-surface-above-ring | s | s | s | s | s |
| stalk-surface-below-ring | s | s | s | s | s |
| stalk-color-above-ring | w | w | w | w | w |
| stalk-color-below-ring | w | w | w | w | w |
| veil-type | p | p | p | p | p |
| veil-color | w | w | w | w | w |
| ring-number | o | o | o | o | o |
| ring-type | p | p | p | p | e |
| spore-print-color | k | n | n | k | n |
| population | s | n | n | s | a |
| habitat | u | g | m | u | g |
shroom_df.isnull().sum()
| class | 0 |
| cap-shape | 0 |
| cap-surface | 0 |
| cap-color | 0 |
| bruises | 0 |
| odor | 0 |
| gill-attachment | 0 |
| gill-spacing | 0 |
| gill-size | 0 |
| gill-color | 0 |
| stalk-shape | 0 |
| stalk-root | 0 |
| stalk-surface-above-ring | 0 |
| stalk-surface-below-ring | 0 |
| stalk-color-above-ring | 0 |
| stalk-color-below-ring | 0 |
| veil-type | 0 |
| veil-color | 0 |
| ring-number | 0 |
| ring-type | 0 |
| spore-print-color | 0 |
| population | 0 |
| habitat | 0 |
| dtype: int64 |
feature_df = shroom_df.describe().transpose().reset_index(
names=['feature']
).sort_values(
'unique', ascending=False
)
| feature | count | unique | top | freq | |
|---|---|---|---|---|---|
| 9 | gill-color | 8124 | 12 | b | 1728 |
| 3 | cap-color | 8124 | 10 | n | 2284 |
| 20 | spore-print-color | 8124 | 9 | w | 2388 |
| 5 | odor | 8124 | 9 | n | 3528 |
| 15 | stalk-color-below-ring | 8124 | 9 | w | 4384 |
| 14 | stalk-color-above-ring | 8124 | 9 | w | 4464 |
| 22 | habitat | 8124 | 7 | d | 3148 |
| 1 | cap-shape | 8124 | 6 | x | 3656 |
| 21 | population | 8124 | 6 | v | 4040 |
| 19 | ring-type | 8124 | 5 | p | 3968 |
| 11 | stalk-root | 8124 | 5 | b | 3776 |
| 12 | stalk-surface-above-ring | 8124 | 4 | s | 5176 |
| 13 | stalk-surface-below-ring | 8124 | 4 | s | 4936 |
| 17 | veil-color | 8124 | 4 | w | 7924 |
| 2 | cap-surface | 8124 | 4 | y | 3244 |
| 18 | ring-number | 8124 | 3 | o | 7488 |
| 10 | stalk-shape | 8124 | 2 | t | 4608 |
| 8 | gill-size | 8124 | 2 | b | 5612 |
| 7 | gill-spacing | 8124 | 2 | c | 6812 |
| 6 | gill-attachment | 8124 | 2 | f | 7914 |
| 4 | bruises | 8124 | 2 | f | 4748 |
| 0 | class | 8124 | 2 | e | 4208 |
| 16 | veil-type | 8124 | 1 | p | 8124 |
plt.figure(figsize=(12,8))
plt.title('Mushroom Features :: Number of unique Features')
sns.barplot(data=feature_df, y='feature', x='unique', orient='h', palette='summer_r')
plt.figure(figsize=(10,4))
plt.title('Mushroom Count :: Editable vs Poisonous')
sns.countplot(data=shroom_df, x='class', palette='seismic_r')
Adaptive Boosting
# remove lable class
X_shroom = shroom_df.drop('class', axis=1)
# make all values numeric
X_shroom = pd.get_dummies(X_shroom, drop_first=True)
y_shroom = shroom_df['class']
# train/test split
X_shroom_train, X_shroom_test, y_shroom_train, y_shroom_test = train_test_split(
X_shroom,
y_shroom,
test_size=0.15,
random_state=42
)
Feature Exploration
# don't try fit a perfect model but only return
# the most important feature for classification
abc_shroom = AdaBoostClassifier(estimator=None, n_estimators=1)
abc_shroom.fit(X_shroom_train,y_shroom_train)
shroom_preds = abc_shroom.predict(X_shroom_test)
print('Accuracy Score: ',accuracy_score(y_shroom_test, shroom_preds, normalize=True).round(4)*100, '%')
# Accuracy Score: 88.35 %
report_shroom = classification_report(y_shroom_test, shroom_preds)
print(report_shroom)
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| e | 0.97 | 0.80 | 0.88 | 637 |
| p | 0.82 | 0.97 | 0.89 | 582 |
| accuracy | 0.88 | 1219 | ||
| macro avg | 0.89 | 0.89 | 0.88 | 1219 |
| weighted avg | 0.90 | 0.88 | 0.88 | 1219 |
conf_mtx_shroom = confusion_matrix(y_shroom_test, shroom_preds)
conf_mtx_shroom_plot = ConfusionMatrixDisplay(
confusion_matrix=conf_mtx_shroom
)
conf_mtx_shroom_plot.plot(cmap='winter_r')
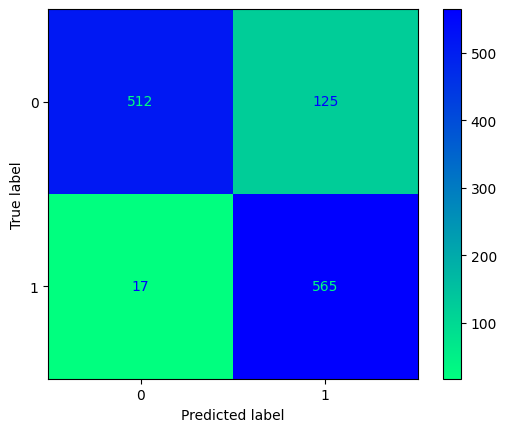
# the model was fit on a single feature and still resulted in a pretty good performance.
# Let's find out what feature was chosen for the classification.
shroom_index = ['importance']
shroom_data_columns = pd.Series(X_shroom.columns)
shroom_importance_array = abc_shroom.feature_importances_
shroom_importance_df = pd.DataFrame(shroom_importance_array, shroom_data_columns, shroom_index)
shroom_importance_df.value_counts()
| importance | count |
|---|---|
| 0.0 | 94 |
| 1.0 | 1 |
| dtype: int64 |
# plot a slice of the dataframe to find the feature
shroom_importance_df_sorted = shroom_importance_df.sort_values(
by='importance',
ascending=True
)
shroom_importance_df_sorted[-5:].plot(
kind='barh',
title='Feature Importance for Mushroom Classification',
figsize=(8,4)
)
The most important feature (as determined by the model) is the odor - in this case a odor of none is the best indicator to classify a poisonous mushroom:
odor: almond = a, anise = l, creosote = c, fishy = y, foul = f, musty = m, none = n, pungent = p, spicy = s
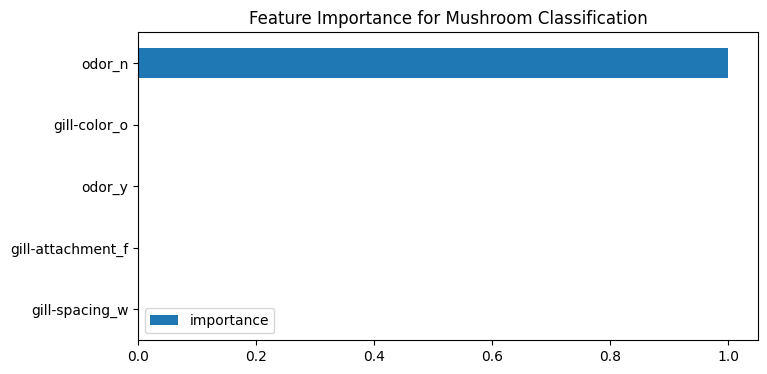
# the mojority of poisonous mushrooms do have an odor
# naking the lack of it a good indicator for an eatable variety
plt.figure(figsize=(12,4))
plt.title('Mushroom Odor vs Class')
sns.countplot(data=shroom_df, x='odor', hue='class', palette='summer')
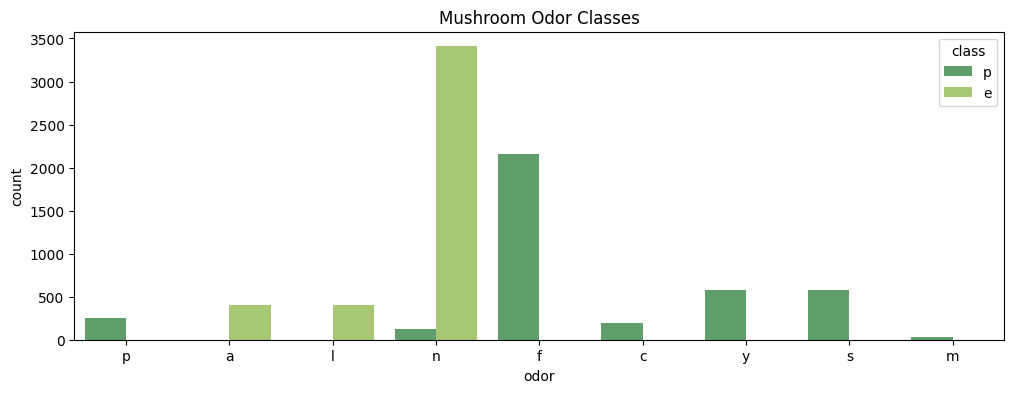
Optimizing Hyperparameters
# find out how many of the 95 features you have
# to add to your model to get a better fit
error_rates = []
for estimators in range(1,96):
model = AdaBoostClassifier(n_estimators=estimators)
model.fit(X_shroom_train,y_shroom_train)
preds = model.predict(X_shroom_test)
err = 1 - accuracy_score(y_shroom_test, preds)
error_rates.append(err)
x_range=range(1,96)
plt.figure(figsize=(10,4))
plt.title('Adaboost Error Rate vs n_estimators')
plt.xlabel('n_estimators')
plt.ylabel('Error Rate')
plt.xticks(np.arange(min(x_range), max(x_range)+1, 3.0))
plt.plot(x_range, error_rates)
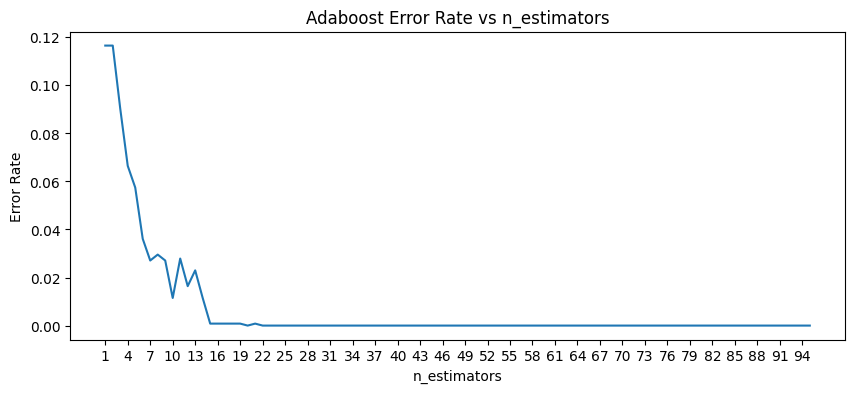
# already after 16 estimators there is no
# visible improvment for the error rate
abc_shroom2 = AdaBoostClassifier(estimator=None, n_estimators=16)
abc_shroom2.fit(X_shroom_train,y_shroom_train)
shroom_preds2 = abc_shroom2.predict(X_shroom_test)
print('Accuracy Score: ',accuracy_score(y_shroom_test, shroom_preds2, normalize=True).round(4)*100, '%')
# Accuracy Score: 99.92 %
report_shroom2 = classification_report(y_shroom_test, shroom_preds2)
print(report_shroom2)
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| e | 1.00 | 1.00 | 1.00 | 637 |
| p | 1.00 | 1.00 | 1.00 | 582 |
| accuracy | 1.00 | 1219 | ||
| macro avg | 1.00 | 1.00 | 1.00 | 1219 |
| weighted avg | 1.00 | 1.00 | 1.00 | 1219 |
conf_mtx_shroom2 = confusion_matrix(y_shroom_test, shroom_preds2)
conf_mtx_shroom_plot2 = ConfusionMatrixDisplay(
confusion_matrix=conf_mtx_shroom2
)
conf_mtx_shroom_plot2.plot(cmap='winter_r')
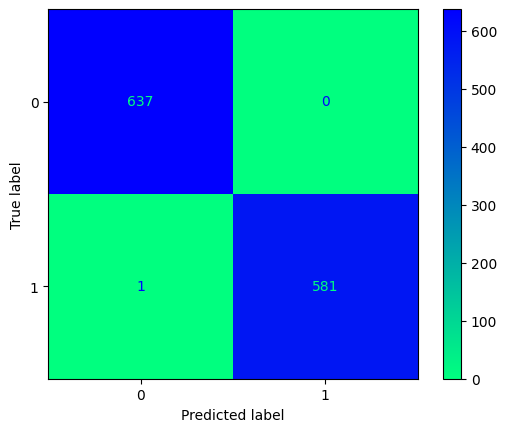
shroom_index = ['importance']
shroom_data_columns = pd.Series(X_shroom.columns)
shroom_importance_array = abc_shroom2.feature_importances_
shroom_importance_df = pd.DataFrame(shroom_importance_array, shroom_data_columns, shroom_index)
shroom_importance_df.value_counts()
# there are 12 features now that are deemed important
| importance | count |
|---|---|
| 0.0000 | 83 |
| 0.0625 | 9 |
| 0.1250 | 2 |
| 0.1875 | 1 |
| dtype: int64 |
shroom_importance_df_sorted = shroom_importance_df.sort_values(
by='importance',
ascending=True
).tail(13)
| importance | |
|---|---|
| gill-size_n | 0.1875 |
| population_v | 0.1250 |
| odor_n | 0.1250 |
| odor_c | 0.0625 |
| stalk-shape_t | 0.0625 |
| spore-print-color_w | 0.0625 |
| population_c | 0.0625 |
| ring-type_p | 0.0625 |
| spore-print-color_r | 0.0625 |
| stalk-surface-above-ring_k | 0.0625 |
| gill-spacing_w | 0.0625 |
| odor_f | 0.0625 |
| stalk-color-below-ring_w | 0.0000 |
plt.figure(figsize=(10,6))
plt.title('Features important to classify poisonous Mushrooms')
sns.barplot(
data=shroom_importance_df_sorted.tail(13),
y=shroom_importance_df_sorted.tail(13).index,
x='importance',
orient='h',
palette='summer'
)
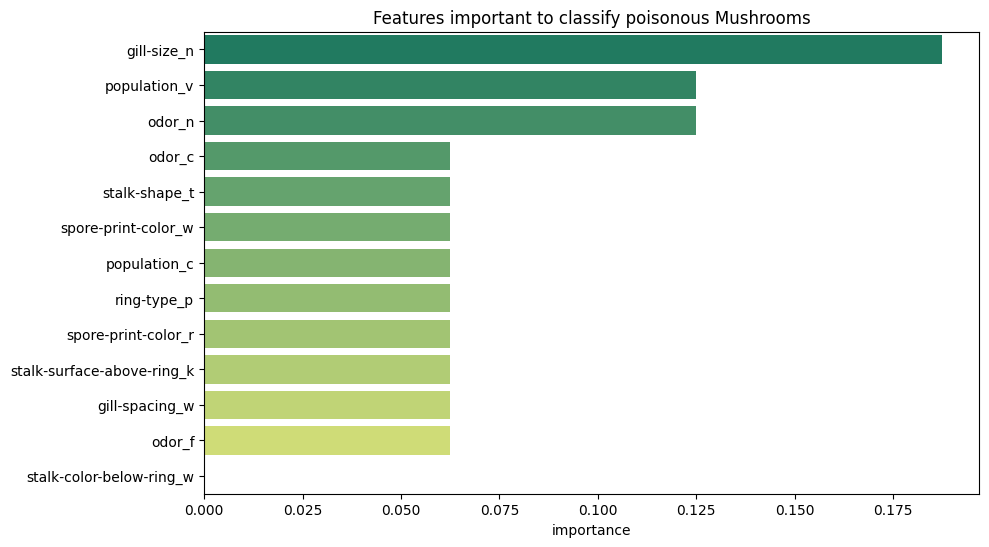
Gradient Boosting
Gridsearch for best Hyperparameter
gb_shroom = GradientBoostingClassifier()
param_grid = \{
'n_estimators': [50, 100, 150],
'learning_rate': [0.05,0.1,0.2],
'max_depth': [2,3,4,5]
\}
shroom_grid = GridSearchCV(gb_shroom, param_grid)
shroom_grid.fit(X_shroom_train, y_shroom_train)
shroom_grid.best_params_
# \{'learning_rate': 0.05, 'max_depth': 4, 'n_estimators': 150\}
shroom_grid_preds = shroom_grid.predict(X_shroom_test)
print('Accuracy Score: ',accuracy_score(y_shroom_test, shroom_grid_preds, normalize=True).round(4)*100, '%')
# Accuracy Score: 100.0 %
report_shroom_grid_preds = classification_report(y_shroom_test, shroom_grid_preds)
print(report_shroom_grid_preds)
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| e | 1.00 | 1.00 | 1.00 | 637 |
| p | 1.00 | 1.00 | 1.00 | 582 |
| accuracy | 1.00 | 1219 | ||
| macro avg | 1.00 | 1.00 | 1.00 | 1219 |
| weighted avg | 1.00 | 1.00 | 1.00 | 1219 |
conf_mtx_shroom_grid = confusion_matrix(y_shroom_test, shroom_grid_preds)
conf_mtx_shroom_grid_plot = ConfusionMatrixDisplay(
confusion_matrix=conf_mtx_shroom_grid
)
conf_mtx_shroom_grid_plot.plot(cmap='winter_r')
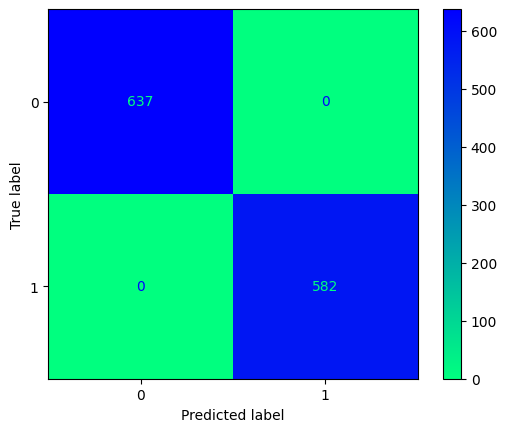
Feature Importance
shroom_feature_importance = shroom_grid.best_estimator_.feature_importances_
feature_importance_df = pd.DataFrame(
index = X_shroom.columns,
data = shroom_feature_importance,
columns = ['importance']
)
# kick all features that have zero importance and sort by importance
feature_importance_df = feature_importance_df[
feature_importance_df['importance'] > 3e-03
].sort_values(
by='importance',
ascending=False
)
plt.figure(figsize=(10,6))
plt.title('Features important to classify poisonous Mushrooms')
sns.barplot(
data=feature_importance_df,
y=feature_importance_df.index,
x='importance',
orient='h',
palette='summer'
)
Supervised Learning - Naive Bayes NLP
Feature Extraction
text = [
'This is a dataset for binary sentiment classification',
'containing substantially more data than previous benchmark datasets',
'We provide a set of 25,000 highly polar movie reviews for training',
'And 25,000 for testing',
'There is additional unlabeled data for use as well',
'Raw text and already processed bag of words formats are provided'
]
CountVectorizer & TfidfTransformer
cv = CountVectorizer(stop_words='english')
cv_sparse_matrix = cv.fit_transform(text)
# <6x30 sparse matrix of type '<class 'numpy.int64'>'
# with 33 stored elements in Compressed Sparse Row format>
print(cv_sparse_matrix.todense())
# [[0 0 0 0 0 1 1 0 0 1 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0]
# [0 0 0 0 1 0 0 1 1 0 1 0 0 0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 0 0]
# [1 1 0 0 0 0 0 0 0 0 0 0 1 1 1 0 0 1 0 0 1 0 1 0 0 0 1 0 0 0]
# [1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0]
# [0 0 1 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0]
# [0 0 0 1 0 0 0 0 0 0 0 1 0 0 0 0 1 0 1 1 0 0 0 0 0 1 0 0 0 1]]
print(cv.vocabulary_)
# \{'dataset': 9, 'binary': 5, 'sentiment': 21, 'classification': 6, 'containing': 7, 'substantially': 23, 'data': 8, 'previous': 15, 'benchmark': 4, 'datasets': 10, 'provide': 17, 'set': 22, '25': 1, '000': 0, 'highly': 12, 'polar': 14, 'movie': 13, 'reviews': 20, 'training': 26, 'testing': 24, 'additional': 2, 'unlabeled': 27, 'use': 28, 'raw': 19, 'text': 25, 'processed': 16, 'bag': 3, 'words': 29, 'formats': 11, 'provided': 18\}
tfidf_trans = TfidfTransformer()
tfidf_trans_results = tfidf_trans.fit_transform(cv_sparse_matrix)
print(tfidf_trans_results.todense())
# [[0. 0. 0. 0. 0. 0.5
# 0.5 0. 0. 0.5 0. 0.
# 0. 0. 0. 0. 0. 0.
# 0. 0. 0. 0.5 0. 0.
# 0. 0. 0. 0. 0. 0. ]
# [0. 0. 0. 0. 0.4198708 0.
# 0. 0.4198708 0.34430007 0. 0.4198708 0.
# 0. 0. 0. 0.4198708 0. 0.
# 0. 0. 0. 0. 0. 0.4198708
# 0. 0. 0. 0. 0. 0. ]
# [0.28386526 0.28386526 0. 0. 0. 0.
# 0. 0. 0. 0. 0. 0.
# 0.3461711 0.3461711 0.3461711 0. 0. 0.3461711
# 0. 0. 0.3461711 0. 0.3461711 0.
# 0. 0. 0.3461711 0. 0. 0. ]
# [0.5355058 0.5355058 0. 0. 0. 0.
# 0. 0. 0. 0. 0. 0.
# 0. 0. 0. 0. 0. 0.
# 0. 0. 0. 0. 0. 0.
# 0.65304446 0. 0. 0. 0. 0. ]
# [0. 0. 0.52182349 0. 0. 0.
# 0. 0. 0.42790272 0. 0. 0.
# 0. 0. 0. 0. 0. 0.
# 0. 0. 0. 0. 0. 0.
# 0. 0. 0. 0.52182349 0.52182349 0. ]
# [0. 0. 0. 0.37796447 0. 0.
# 0. 0. 0. 0. 0. 0.37796447
# 0. 0. 0. 0. 0.37796447 0.
# 0.37796447 0.37796447 0. 0. 0. 0.
# 0. 0.37796447 0. 0. 0. 0.37796447]]
TfidfVectorizer
tfidf_vec = TfidfVectorizer(
lowercase=True,
analyzer='word',
stop_words='english'
)
tfidf_vec_results = tfidf_vec.fit_transform(text)
# <6x30 sparse matrix of type '<class 'numpy.float64'>'
# with 33 stored elements in Compressed Sparse Row format>
print(tfidf_trans_results == tfidf_vec_results)
# True
Dataset Exploration
!wget https://raw.githubusercontent.com/kunal-lalwani/Twitter-US-Airlines-Sentiment-Analysis/master/Tweets.csv -P datasets
tweet_df = pd.read_csv('datasets/Tweets.csv')
tweet_df.head(3).transpose()
| 0 | 1 | 2 | |
|---|---|---|---|
| tweet_id | 570306133677760513 | 570301130888122368 | 570301083672813571 |
| airline_sentiment | neutral | positive | neutral |
| airline_sentiment_confidence | 1.0 | 0.3486 | 0.6837 |
| negativereason | NaN | NaN | NaN |
| negativereason_confidence | NaN | 0.0 | NaN |
| airline | Virgin America | Virgin America | Virgin America |
| airline_sentiment_gold | NaN | NaN | NaN |
| name | cairdin | jnardino | yvonnalynn |
| negativereason_gold | NaN | NaN | NaN |
| retweet_count | 0 | 0 | 0 |
| text | @VirginAmerica What @dhepburn said. | @VirginAmerica plus you've added commercials t... | @VirginAmerica I didn't today... Must mean I n... |
| tweet_coord | NaN | NaN | NaN |
| tweet_created | 2015-02-24 11:35:52 -0800 | 2015-02-24 11:15:59 -0800 | 2015-02-24 11:15:48 -0800 |
| tweet_location | NaN | NaN | Lets Play |
| user_timezone | Eastern Time (US & Canada) | Pacific Time (US & Canada) | Central Time (US & Canada) |
plt.figure(figsize=(12,5))
plt.title('Tweet Sentiment Classification by Airline')
sns.countplot(
data=tweet_df,
x='airline',
hue='airline_sentiment',
palette='cool'
)
plt.savefig('assets/Scikit_Learn_56.webp', bbox_inches='tight')
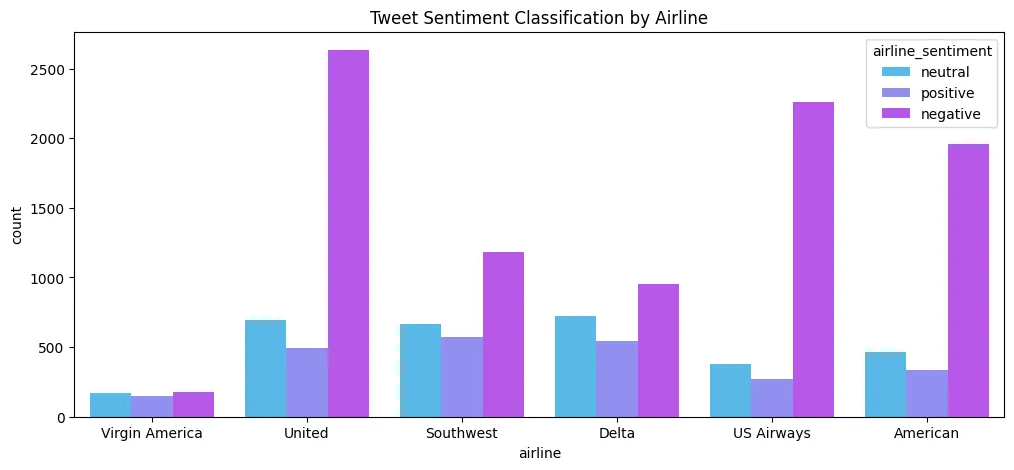
plt.figure(figsize=(12,6))
plt.title('Tweet Sentiment Classification with negative Reason')
sns.countplot(
data=tweet_df,
x='airline',
hue='negativereason',
palette='cool'
)
plt.savefig('assets/Scikit_Learn_57.webp', bbox_inches='tight')
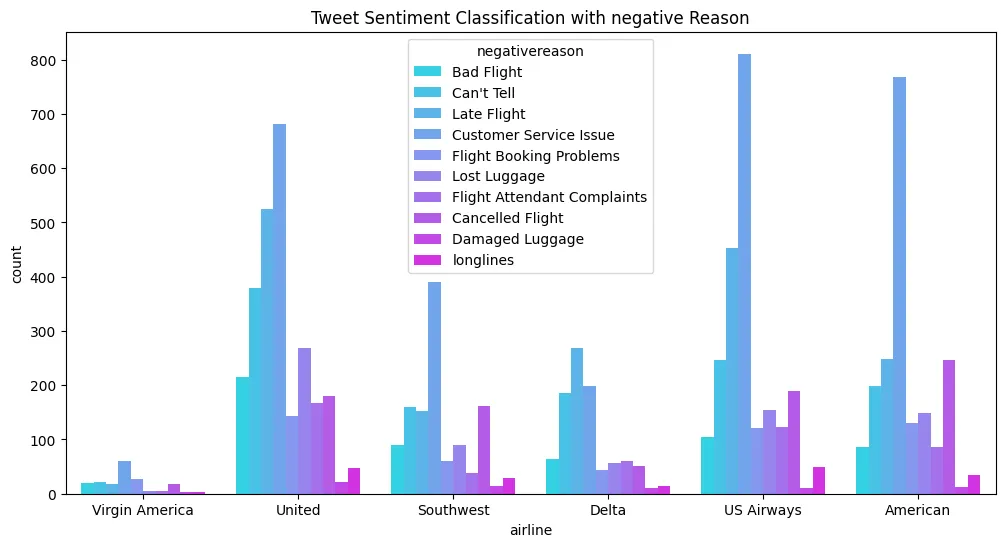
Data Preprocessing
tweet_data = tweet_df[['airline_sentiment', 'text']]
X_tweet = tweet_data['text']
y_tweet = tweet_data['airline_sentiment']
# train/ test split
X_tweet_train, X_tweet_test, y_tweet_train, y_tweet_test = train_test_split(
X_tweet,
y_tweet,
test_size=0.2,
random_state=42
)
TFIDF Vectorizer
tfidf_tweet_vec = TfidfVectorizer(
lowercase=True,
analyzer='word',
stop_words='english'
)
X_tweet_tfidf_train = tfidf_tweet_vec.fit_transform(X_tweet_train)
# <11712x12987 sparse matrix of type '<class 'numpy.float64'>'
# with 106745 stored elements in Compressed Sparse Row format>
X_tweet_tfidf_test = tfidf_tweet_vec.transform(X_tweet_test)
Model Comparison
# report helper function
def report(model):
preds = model.predict(X_tweet_tfidf_test)
print(classification_report(y_tweet_test, preds))
conf_mtx = confusion_matrix(y_tweet_test, preds)
conf_mtx_plot = ConfusionMatrixDisplay(
confusion_matrix=conf_mtx
)
conf_mtx_plot.plot(cmap='plasma')
logreg_tweet = LogisticRegression(max_iter=1000)
logreg_tweet.fit(X_tweet_tfidf_train, y_tweet_train)
report(logreg_tweet)
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| negative | 0.82 | 0.93 | 0.88 | 1889 |
| neutral | 0.66 | 0.48 | 0.56 | 580 |
| positive | 0.79 | 0.63 | 0.70 | 459 |
| accuracy | 0.80 | 2928 | ||
| macro avg | 0.76 | 0.68 | 0.71 | 2928 |
| weighted avg | 0.79 | 0.80 | 0.78 | 2928 |
rbf_svc_tweet = svm.SVC()
rbf_svc_tweet.fit(X_tweet_tfidf_train, y_tweet_train)
report(rbf_svc_tweet)
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| negative | 0.81 | 0.95 | 0.87 | 1889 |
| neutral | 0.68 | 0.42 | 0.52 | 580 |
| positive | 0.80 | 0.61 | 0.69 | 459 |
| accuracy | 0.79 | 2928 | ||
| macro avg | 0.76 | 0.66 | 0.69 | 2928 |
| weighted avg | 0.78 | 0.79 | 0.77 | 2928 |
linear_svc_tweet = svm.LinearSVC()
linear_svc_tweet.fit(X_tweet_tfidf_train, y_tweet_train)
report(linear_svc_tweet)
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| negative | 0.85 | 0.91 | 0.88 | 1889 |
| neutral | 0.64 | 0.54 | 0.58 | 580 |
| positive | 0.76 | 0.67 | 0.71 | 459 |
| accuracy | 0.80 | 2928 | ||
| macro avg | 0.75 | 0.71 | 0.72 | 2928 |
| weighted avg | 0.79 | 0.80 | 0.79 | 2928 |
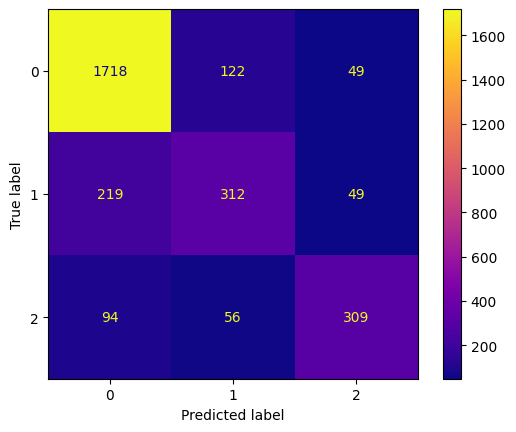
nb_tweets = MultinomialNB()
nb_tweets.fit(X_tweet_tfidf_train, y_tweet_train)
report(nb_tweets)
# The Naive Bayes classifies almost all tweets as negative
# which means it does well with searching neg tweets
# but ends up classifying a lot neutral and pos tweets as neg
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| negative | 0.69 | 0.99 | 0.81 | 1889 |
| neutral | 0.75 | 0.15 | 0.25 | 580 |
| positive | 0.94 | 0.18 | 0.31 | 459 |
| accuracy | 0.70 | 2928 | ||
| macro avg | 0.79 | 0.44 | 0.46 | 2928 |
| weighted avg | 0.74 | 0.70 | 0.62 | 2928 |
Model Deployment
# building a pipeline to ingest new tweets with the best performing model
pipe = Pipeline(
[
('tfidf', TfidfVectorizer()),
('svc', svm.SVC())
]
)
# before deployment retrain on entire dataset
pipe.fit(X_tweet, y_tweet)
# test prediction
print(pipe.predict([
'good flight',
'terrible service',
'too late',
'ok flight',
'Thank you'
]))
# ['positive' 'negative' 'negative' 'neutral' 'positive']
Text Classification
IMDB Dataset of 50K Movie Reviews https://ai.stanford.edu/~amaas/data/sentiment/
Data Exploration
imdb_df = pd.read_csv('datasets/moviereviews.csv')
imdb_df.head()
| label | review | |
|---|---|---|
| 0 | neg | how do films like mouse hunt get into theatres... |
| 1 | neg | some talented actresses are blessed with a dem... |
| 2 | pos | this has been an extraordinary year for austra... |
| 3 | pos | according to hollywood movies made in last few... |
| 4 | neg | my first press screening of 1998 and already i... |
imdb_df.info()
# <class 'pandas.core.frame.DataFrame'>
# RangeIndex: 2000 entries, 0 to 1999
# Data columns (total 2 columns):
# # Column Non-Null Count Dtype
# --- ------ -------------- -----
# 0 label 2000 non-null object
# 1 review 1965 non-null object
# dtypes: object(2)
# memory usage: 31.4+ KB
# find missing
imdb_df.isnull().sum()
# label 0
# review 35
# dtype: int64
# drop missing
imdb_df = imdb_df.dropna(axis=0)
imdb_df.isnull().sum()
# label 0
# review 0
# dtype: int64
# make sure there a no empty string reviews
# (imdb_df['review'] == ' ').sum()
imdb_df['review'].str.isspace().sum()
# 27
# remove empty string reviews
imdb_df = imdb_df[~imdb_df['review'].str.isspace()]
imdb_df = imdb_df[imdb_df['review'] != '']
imdb_df['review'].str.isspace().sum()
# 0
# is the dataset balanced
imdb_df['label'].value_counts()
# neg 969
# pos 969
# Name: label, dtype: int64
Top 30 Features by Label
# find top 20 words in negative reviews
imdb_neg_df = imdb_df[imdb_df['label'] == 'neg']
count_vectorizer = CountVectorizer(analyzer='word', stop_words='english')
bag_of_words = count_vectorizer.fit_transform(imdb_neg_df['review'])
sum_words = bag_of_words.sum(axis=0)
words_freq = [
(word, sum_words[0, idx]) for word, idx in count_vectorizer.vocabulary_.items()
]
words_freq =sorted(words_freq, key = lambda x: x[1], reverse=True)
x, y = zip(*words_freq[:30])
plt.figure(figsize=(12,5))
plt.bar(x,y)
plt.xticks(rotation=90)
plt.title('Top30 Words used in Negative Reviews')
plt.savefig('assets/Scikit_Learn_62.webp', bbox_inches='tight')
# find top 20 words in positive reviews
imdb_pos_df = imdb_df[imdb_df['label'] != 'neg']
count_vectorizer = CountVectorizer(analyzer='word', stop_words='english')
bag_of_words = count_vectorizer.fit_transform(imdb_pos_df['review'])
sum_words = bag_of_words.sum(axis=0)
words_freq = [
(word, sum_words[0, idx]) for word, idx in count_vectorizer.vocabulary_.items()
]
words_freq =sorted(words_freq, key = lambda x: x[1], reverse=True)
x, y = zip(*words_freq[:30])
plt.figure(figsize=(12,5))
plt.bar(x,y)
plt.xticks(rotation=90)
plt.title('Top30 Words used in Positive Reviews')
plt.savefig('assets/Scikit_Learn_63.webp', bbox_inches='tight')
Data Preprocessing
X_rev = imdb_df['review']
y_rev = imdb_df['label']
# train/ test split
X_rev_train, X_rev_test, y_rev_train, y_rev_test = train_test_split(
X_rev,
y_rev,
test_size=0.2,
random_state=42
)
tfidf_rev_vec = TfidfVectorizer(
lowercase=True,
analyzer='word',
stop_words='english'
)
X_rev_tfidf_train = tfidf_rev_vec.fit_transform(X_rev_train)
X_rev_tfidf_test = tfidf_rev_vec.transform(X_rev_test)
Model Training
nb_rev = MultinomialNB()
nb_rev.fit(X_rev_tfidf_train, y_rev_train)
preds = nb_rev.predict(X_rev_tfidf_test)
print(classification_report(y_rev_test, preds))
| precision | recall | f1-score | support | |
|---|---|---|---|---|
| neg | 0.79 | 0.88 | 0.83 | 188 |
| pos | 0.87 | 0.78 | 0.82 | 200 |
| accuracy | 0.82 | 388 | ||
| macro avg | 0.83 | 0.83 | 0.82 | 388 |
| weighted avg | 0.83 | 0.82 | 0.82 | 388 |
conf_mtx = confusion_matrix(y_rev_test, preds)
conf_mtx_plot = ConfusionMatrixDisplay(
confusion_matrix=conf_mtx
)
conf_mtx_plot.plot(cmap='plasma')
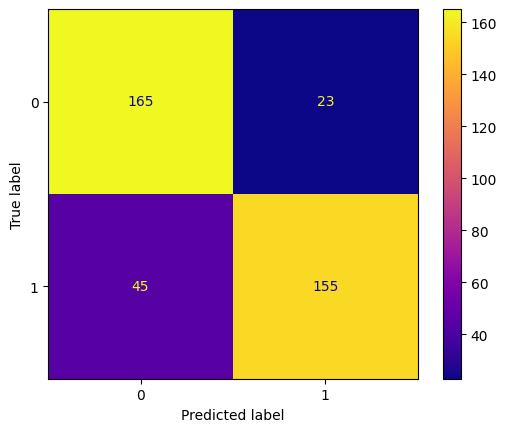
Unsupervised Learning - KMeans Clustering
Dataset Exploration
!wget https://github.com/selva86/datasets/raw/master/bank-full.csv -P datasets
bank_df = pd.read_csv('datasets/bank-full.csv', sep=';')
bank_df.head(5).transpose()
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| age | 56 | 57 | 37 | 40 | 56 |
| job | housemaid | services | services | admin. | services |
| marital | married | married | married | married | married |
| education | basic.4y | high.school | high.school | basic.6y | high.school |
| default | no | unknown | no | no | no |
| housing | no | no | yes | no | no |
| loan | no | no | no | no | yes |
| contact | telephone | telephone | telephone | telephone | telephone |
| month | may | may | may | may | may |
| day_of_week | mon | mon | mon | mon | mon |
| duration | 261 | 149 | 226 | 151 | 307 |
| campaign | 1 | 1 | 1 | 1 | 1 |
| pdays | 999 | 999 | 999 | 999 | 999 |
| previous | 0 | 0 | 0 | 0 | 0 |
| poutcome | nonexistent | nonexistent | nonexistent | nonexistent | nonexistent |
| emp.var.rate | 1.1 | 1.1 | 1.1 | 1.1 | 1.1 |
| cons.price.idx | 93.994 | 93.994 | 93.994 | 93.994 | 93.994 |
| cons.conf.idx | -36.4 | -36.4 | -36.4 | -36.4 | -36.4 |
| euribor3m | 4.857 | 4.857 | 4.857 | 4.857 | 4.857 |
| nr.employed | 5191.0 | 5191.0 | 5191.0 | 5191.0 | 5191.0 |
| y | no | no | no | no | no |
bank_df.describe()
| age | duration | campaign | pdays | previous | emp.var.rate | cons.price.idx | cons.conf.idx | euribor3m | nr.employed | |
|---|---|---|---|---|---|---|---|---|---|---|
| count | 41188.00000 | 41188.000000 | 41188.000000 | 41188.000000 | 41188.000000 | 41188.000000 | 41188.000000 | 41188.000000 | 41188.000000 | 41188.000000 |
| mean | 40.02406 | 258.285010 | 2.567593 | 962.475454 | 0.172963 | 0.081886 | 93.575664 | -40.502600 | 3.621291 | 5167.035911 |
| std | 10.42125 | 259.279249 | 2.770014 | 186.910907 | 0.494901 | 1.570960 | 0.578840 | 4.628198 | 1.734447 | 72.251528 |
| min | 17.00000 | 0.000000 | 1.000000 | 0.000000 | 0.000000 | -3.400000 | 92.201000 | -50.800000 | 0.634000 | 4963.600000 |
| 25% | 32.00000 | 102.000000 | 1.000000 | 999.000000 | 0.000000 | -1.800000 | 93.075000 | -42.700000 | 1.344000 | 5099.100000 |
| 50% | 38.00000 | 180.000000 | 2.000000 | 999.000000 | 0.000000 | 1.100000 | 93.749000 | -41.800000 | 4.857000 | 5191.000000 |
| 75% | 47.00000 | 319.000000 | 3.000000 | 999.000000 | 0.000000 | 1.400000 | 93.994000 | -36.400000 | 4.961000 | 5228.100000 |
| max | 98.00000 | 4918.000000 | 56.000000 | 999.000000 | 7.000000 | 1.400000 | 94.767000 | -26.900000 | 5.045000 | 5228.100000 |
plt.figure(figsize=(12, 5))
plt.title('Age Distribution by Marital Status')
sns.histplot(
data=bank_df,
x='age',
bins=50,
hue='marital',
palette='winter',
kde=True
)
plt.savefig('assets/Scikit_Learn_65.webp', bbox_inches='tight')
plt.figure(figsize=(12, 5))
plt.title('Age Distribution by Loan Status')
sns.histplot(
data=bank_df,
x='age',
bins=50,
hue='loan',
palette='winter',
kde=True
)
plt.savefig('assets/Scikit_Learn_66.webp', bbox_inches='tight')
# remove columns with `pday`s = 999 (placeholder for never)
plt.figure(figsize=(12, 5))
plt.title('Distribution of Days Since Last Contacted by Loan Status')
sns.histplot(
data=bank_df[bank_df['pdays'] != 999],
x='pdays',
hue='loan',
palette='winter',
kde=True
)
plt.savefig('assets/Scikit_Learn_67.webp', bbox_inches='tight')
# Create call duration in minutes column
bank_df['duration_minutes'] = bank_df['duration'].apply(lambda x: x/60).round(1)
plt.figure(figsize=(12, 5))
plt.title('Distribution Contact Duration by Contact Type')
plt.xlim(0,20)
sns.histplot(
data=bank_df,
x='duration_minutes',
hue='contact',
palette='winter',
kde=True
)
plt.savefig('assets/Scikit_Learn_68.webp', bbox_inches='tight')
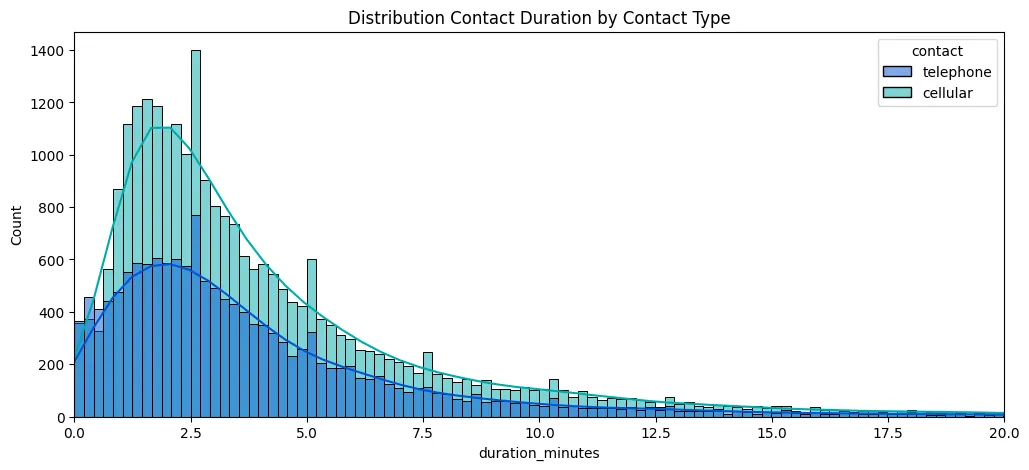
plt.figure(figsize=(16, 5))
plt.title('Customer Jobs Countplot by Loan Defaults')
sns.countplot(
data=bank_df,
x='job',
order=bank_df['job'].value_counts().index,
palette='winter',
hue='default'
)
plt.savefig('assets/Scikit_Learn_69.webp', bbox_inches='tight')
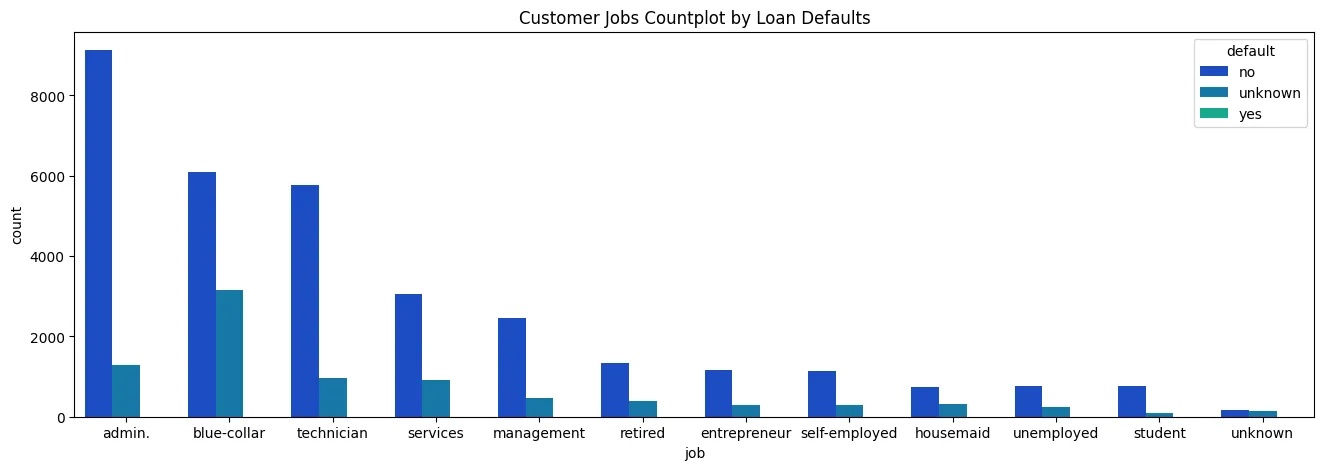
plt.figure(figsize=(16, 5))
plt.title('Customer Education Countplot by Loan Defaults')
sns.countplot(
data=bank_df,
x='education',
order=bank_df['education'].value_counts().index,
palette='winter',
hue='default'
)
plt.savefig('assets/Scikit_Learn_70.webp', bbox_inches='tight')
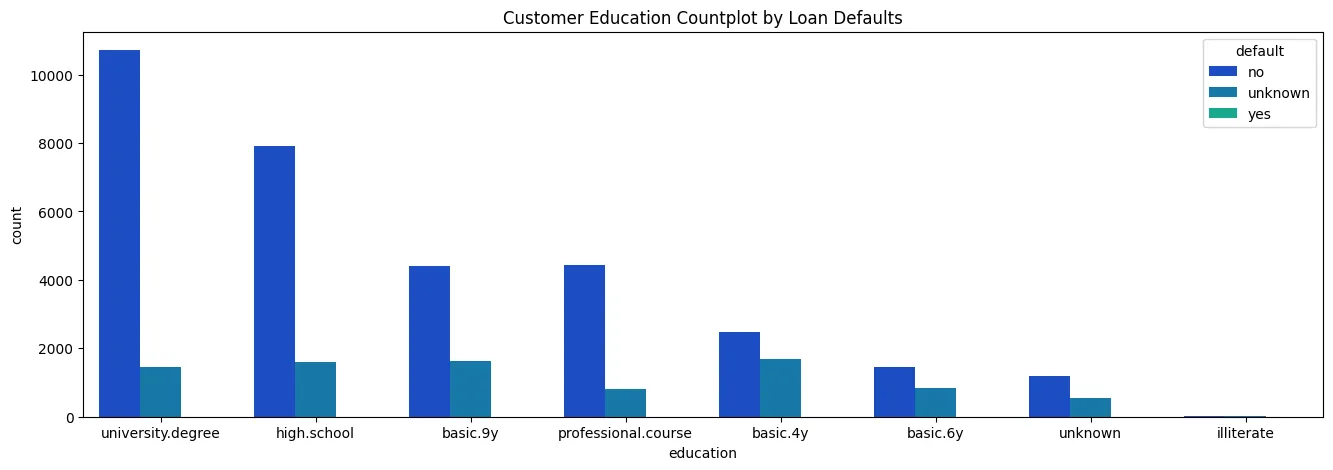
sns.pairplot(
data=bank_df,
hue='marital',
palette='winter'
)
plt.savefig('assets/Scikit_Learn_71.webp', bbox_inches='tight')
Dataset Preprocessing
# encode categorical features
X_bank = pd.get_dummies(bank_df)
# normalize data
bank_scaler = StandardScaler()
X_bank_scaled = bank_scaler.fit_transform(X_bank)
Model Training
bank_model = KMeans(
n_clusters=2,
n_init='auto',
random_state=42
)
# fit to find cluster centers and predict what center every feature belongs to
bank_cluster_labels = bank_model.fit_predict(X_bank_scaled)
# add predicted label to source dataframe
X_bank['Cluster'] = bank_cluster_labels
X_bank['Cluster'].value_counts()
# 0 26871
# 1 14317
# Name: Cluster, dtype: int64
# How do the feature correlate with the predicted labels
label_corr = X_bank.corr()['Cluster']
print(label_corr.iloc[:-1].sort_values())
plt.figure(figsize=(10,14))
label_corr.iloc[:-1].sort_values().plot(kind='barh')
plt.title('Feature Importance')
plt.savefig('assets/Scikit_Learn_72.webp', bbox_inches='tight')
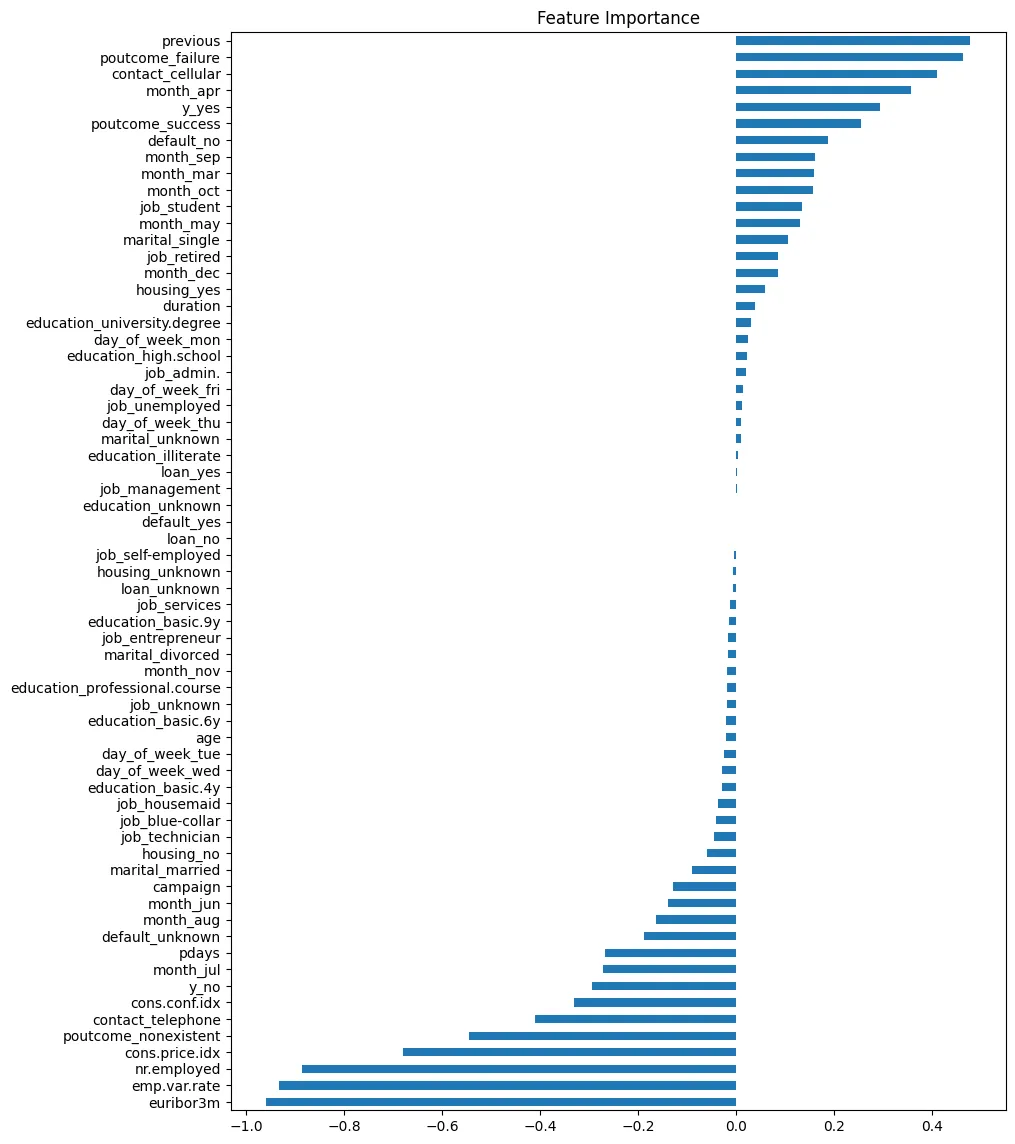
Choosing a K Value
# visualize the sum distance of your datapoints to the
# predicted cluster centers as a function of number of clusters
sum_squared_distance = []
for k in range(2,20):
model = KMeans(n_clusters=k, n_init='auto')
model.fit(X_bank_scaled)
sum_squared_distance.append(model.inertia_)
plt.figure(figsize=(10,5))
plt.title('SSD as a Function of Number of Cluster')
plt.plot(range(2,20), sum_squared_distance, 'o--')
plt.savefig('assets/Scikit_Learn_73.webp', bbox_inches='tight')
plt.figure(figsize=(10,5))
plt.title('Difference in SSD as a Function of Number of Clusters')
pd.Series(sum_squared_distance).diff().plot(kind='bar')
plt.savefig('assets/Scikit_Learn_74.webp', bbox_inches='tight')
There are two 'elbows' - one between k=5-6 (behold the 0-index in Pandas!) and the second one between k=14-15. Both of them are potential good values for the number of cluster
k.
Re-fitting the Model
bank_model = KMeans(
n_clusters=6,
n_init='auto',
random_state=42
)
# fit to find cluster centers and predict what center every feature belongs to
bank_cluster_labels = bank_model.fit_predict(X_bank_scaled)
# add predicted label to source dataframe
X_bank['Cluster'] = bank_cluster_labels
X_bank['Cluster'].value_counts()
# 5 10713
# 0 10663
# 1 8164
# 3 5566
# 4 3322
# 2 2760
# Name: Cluster, dtype: int64
Example 1 : Color Quantization
img_array = mpimg.imread('assets/gz.jpg')
img_array.shape
# (325, 640, 3)
plt.imshow(img_array)
plt.title('Original Image')
plt.savefig('assets/Scikit_Learn_75.webp', bbox_inches='tight')

# flatten the image from 3 to 2 dimensions
(height, width, colour) = img_array.shape
img_array2d = img_array.reshape(height*width,colour)
img_array2d.shape
# (208000, 3)
# reduce colour space to 6 clusters
colour_model = KMeans(n_clusters=6, n_init='auto')
colour_labels = colour_model.fit_predict(img_array2d)
# get rgb value for each of the 6 cluster centers
rgb_colours = colour_model.cluster_centers_.round(0).astype(int)
rgb_colours
# array([[186, 111, 58],
# [ 31, 11, 16],
# [135, 72, 46],
# [236, 157, 73],
# [ 81, 40, 34],
# [252, 199, 125]])
# assign these rgb values to each pixel within the cluster
# and reshape to original 3d array
quantized_image = np.reshape(rgb_colours[colour_labels],(height,width,colour))
plt.imshow(quantized_image)
plt.title('Quantized Image')
plt.savefig('assets/Scikit_Learn_76.webp', bbox_inches='tight')

Example 2 : Country Clustering
Dataset Exploration
!wget https://github.com/priyansh21112002/CIA-Country-Description/raw/main/CIA_Country_Facts.csv -P datasets
country_df = pd.read_csv('datasets/CIA_Country_Facts.csv')
country_df.head(5).transpose()
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| Country | Afghanistan | Albania | Algeria | American Samoa | Andorra |
| Region | ASIA (EX. NEAR EAST) | EASTERN EUROPE | NORTHERN AFRICA | OCEANIA | WESTERN EUROPE |
| Population | 31056997 | 3581655 | 32930091 | 57794 | 71201 |
| Area (sq. mi.) | 647500 | 28748 | 2381740 | 199 | 468 |
| Pop. Density (per sq. mi.) | 48.0 | 124.6 | 13.8 | 290.4 | 152.1 |
| Coastline (coast/area ratio) | 0.0 | 1.26 | 0.04 | 58.29 | 0.0 |
| Net migration | 23.06 | -4.93 | -0.39 | -20.71 | 6.6 |
| Infant mortality (per 1000 births) | 163.07 | 21.52 | 31.0 | 9.27 | 4.05 |
| GDP ($ per capita) | 700.0 | 4500.0 | 6000.0 | 8000.0 | 19000.0 |
| Literacy (%) | 36.0 | 86.5 | 70.0 | 97.0 | 100.0 |
| Phones (per 1000) | 3.2 | 71.2 | 78.1 | 259.5 | 497.2 |
| Arable (%) | 12.13 | 21.09 | 3.22 | 10.0 | 2.22 |
| Crops (%) | 0.22 | 4.42 | 0.25 | 15.0 | 0.0 |
| Other (%) | 87.65 | 74.49 | 96.53 | 75.0 | 97.78 |
| Climate | 1.0 | 3.0 | 1.0 | 2.0 | 3.0 |
| Birthrate | 46.6 | 15.11 | 17.14 | 22.46 | 8.71 |
| Deathrate | 20.34 | 5.22 | 4.61 | 3.27 | 6.25 |
| Agriculture | 0.38 | 0.232 | 0.101 | NaN | NaN |
| Industry | 0.24 | 0.188 | 0.6 | NaN | NaN |
| Service | 0.38 | 0.579 | 0.298 | NaN | NaN |
fig, axes = plt.subplots(figsize=(10,5), nrows=1, ncols=2)
plt.suptitle('Country Population Histogram')
axes[0].set_xlabel('Population')
axes[0].set_ylabel('Frequency')
axes[0].hist(
x=country_df['Population'],
range=None,
density=True,
histtype='bar',
orientation='vertical',
color='dodgerblue'
)
axes[1].set_xlabel('Population (<100Mio)')
axes[1].set_ylabel('Frequency')
axes[1].hist(
x=country_df['Population'],
range=[0, 1e8],
density=True,
histtype='bar',
orientation='vertical',
color='fuchsia'
)
plt.savefig('assets/Scikit_Learn_77.webp', bbox_inches='tight')
plt.figure(figsize=(12, 5))
plt.title('GDP ($ per capita) by Region')
sns.barplot(
data=country_df,
y='Region',
x='GDP ($ per capita)',
estimator=np.mean,
errorbar='sd',
orient='h',
palette='cool'
)
plt.savefig('assets/Scikit_Learn_78.webp', bbox_inches='tight')
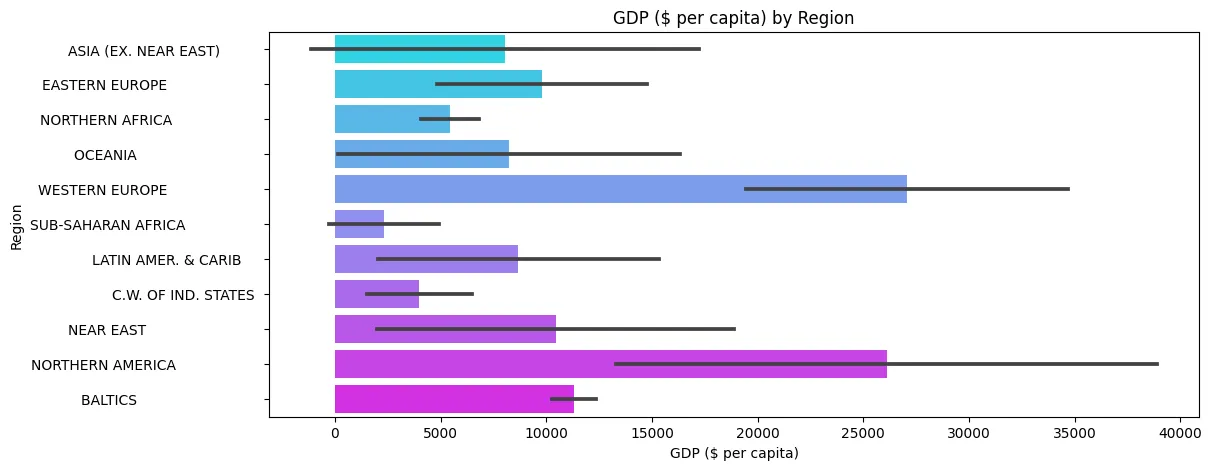
plt.figure(figsize=(10, 6))
sns.scatterplot(
y='Phones (per 1000)',
x='GDP ($ per capita)',
data=country_df,
hue='Region',
palette='cool',
).set_title('GDP ($ per capita) vs. Phones (per 1000)')
plt.savefig('assets/Scikit_Learn_79.webp', bbox_inches='tight')
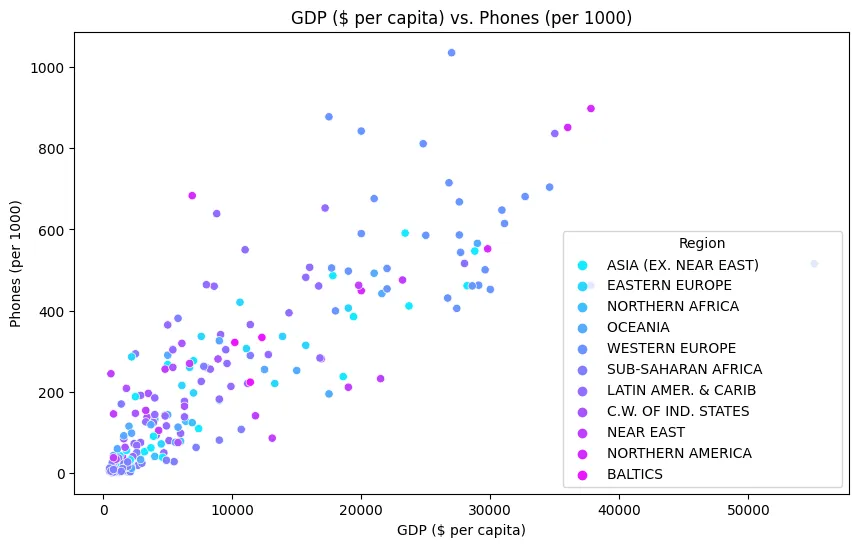
plt.figure(figsize=(10, 6))
sns.scatterplot(
y='Literacy (%)',
x='GDP ($ per capita)',
data=country_df,
hue='Region',
palette='cool',
).set_title('GDP ($ per capita) vs. Literacy (%)')
plt.savefig('assets/Scikit_Learn_80.webp', bbox_inches='tight')
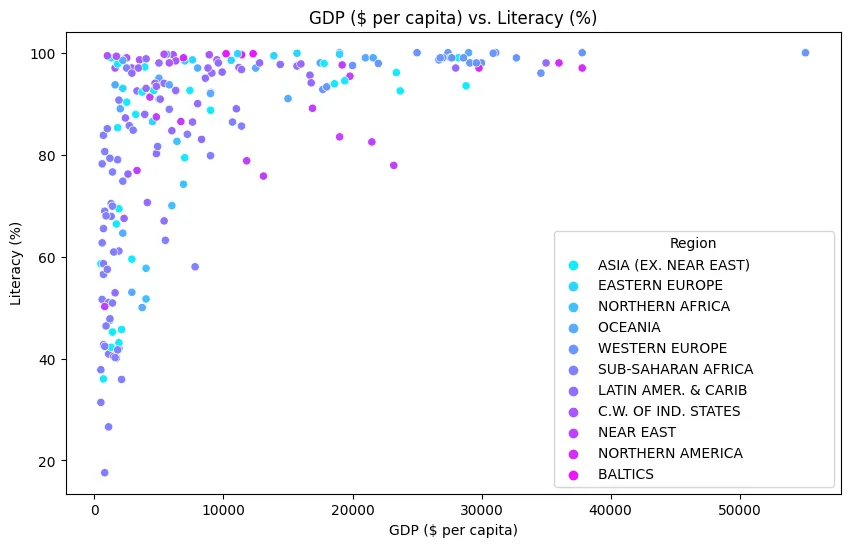
plt.figure(figsize=(20, 12), dpi=200)
plt.title('Correlation Heatmap CIA Country Dataset')
sns.heatmap(
country_df.corr(numeric_only=True),
linewidth=0.5,
cmap='seismic',
annot=True
)
plt.savefig('assets/Scikit_Learn_81.webp', bbox_inches='tight')
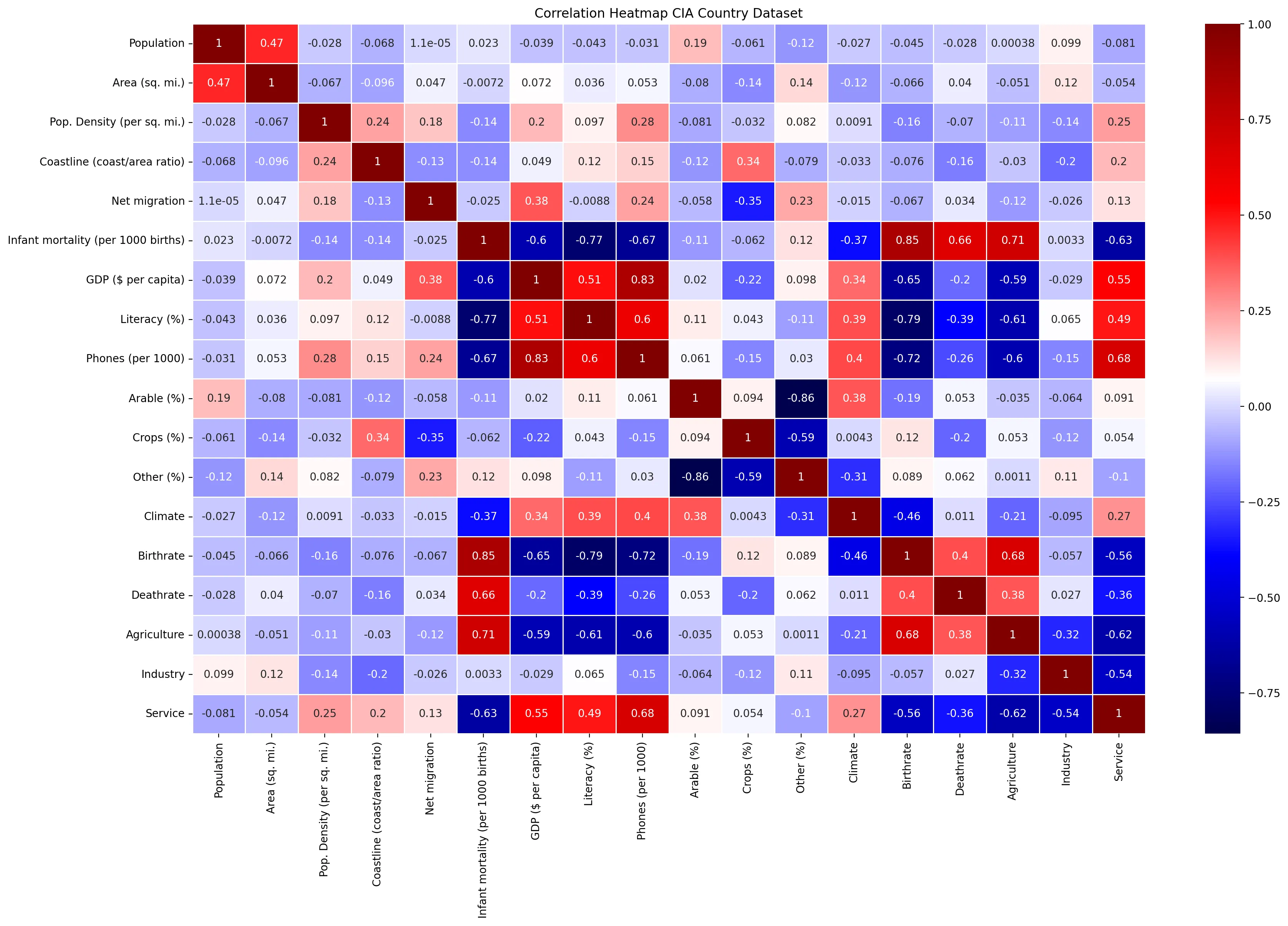
plt.figure(figsize=(20, 12), dpi=200)
sns.clustermap(
country_df.corr(numeric_only=True),
linewidth=0.5,
cmap='seismic',
annot=False,
col_cluster=False
)
plt.savefig('assets/Scikit_Learn_82.webp', bbox_inches='tight')
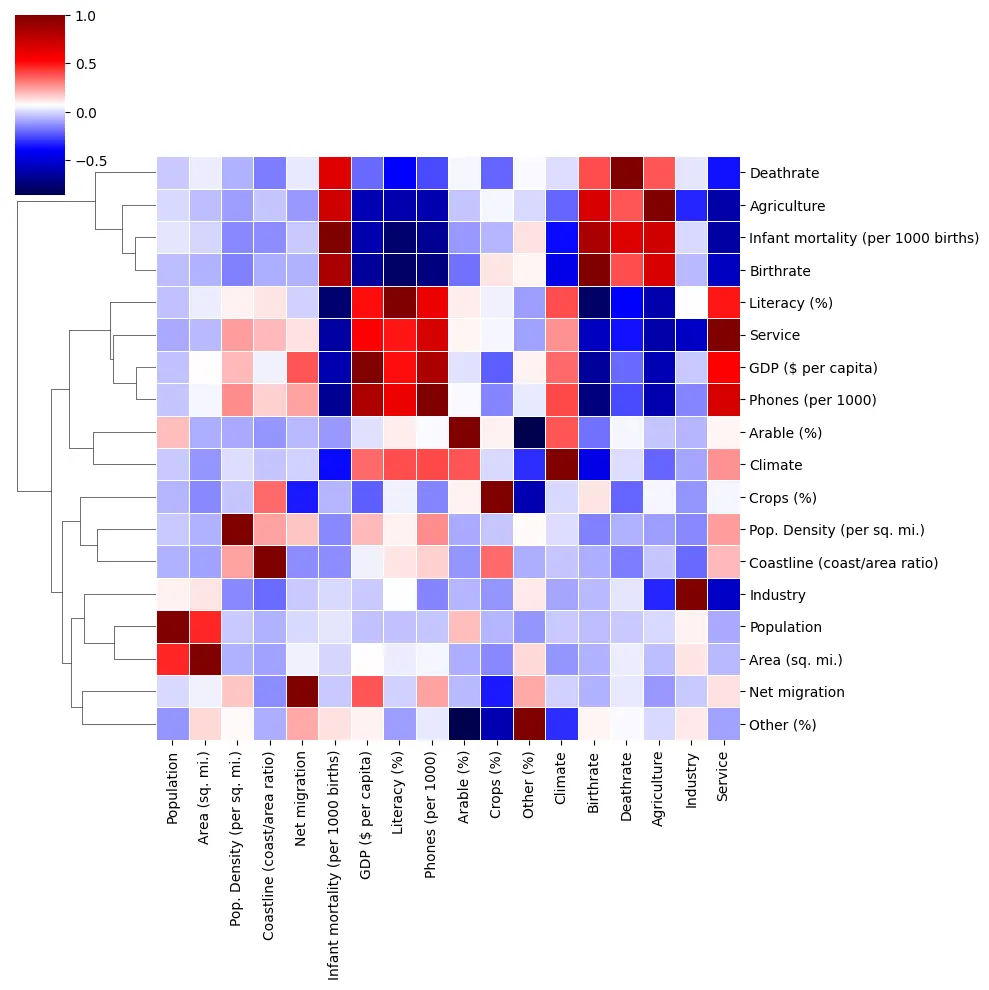
Dataset Preprocessing
# find columns with missing values
country_df.isnull().sum()
| Country | 0 |
| Region | 0 |
| Population | 0 |
| Area (sq. mi.) | 0 |
| Pop. Density (per sq. mi.) | 0 |
| Coastline (coast/area ratio) | 0 |
| Net migration | 3 |
| Infant mortality (per 1000 births) | 3 |
| GDP ($ per capita) | 1 |
| Literacy (%) | 18 |
| Phones (per 1000) | 4 |
| Arable (%) | 2 |
| Crops (%) | 2 |
| Other (%) | 2 |
| Climate | 22 |
| Birthrate | 3 |
| Deathrate | 4 |
| Agriculture | 15 |
| Industry | 16 |
| Service | 15 |
| dtype: int64 |
# what countries don't have an agriculture value
country_df[pd.isnull(country_df['Agriculture'])]['Country']
# all countries without agriculture data will not have a
# whole lot of agriculture output. The same is true for 'Industry'
# and 'Service' These values can be set to zero:
| 3 | American Samoa |
| 4 | Andorra |
| 78 | Gibraltar |
| 80 | Greenland |
| 83 | Guam |
| 134 | Mayotte |
| 140 | Montserrat |
| 144 | Nauru |
| 153 | N. Mariana Islands |
| 171 | Saint Helena |
| 174 | St Pierre & Miquelon |
| 177 | San Marino |
| 208 | Turks & Caicos Is |
| 221 | Wallis and Futuna |
| 223 | Western Sahara |
| Name: Country, dtype: object |
# set missing values to zero for Agriculture, Industry and Service
# define what default values you want to fill
values = \{
"Agriculture": 0,
"Industry": 0,
"Service": 0,
\}
# and replace missing with values
country_df = country_df.fillna(value=values)
# another datapoint that is often missing is climate
# the climate can be estimated by countries in the same Region
country_df[pd.isnull(country_df['Climate'])][['Country', 'Region', 'Climate']]
| Country | Region | Climate | |
|---|---|---|---|
| 5 | Angola | SUB-SAHARAN AFRICA | NaN |
| 36 | Canada | NORTHERN AMERICA | NaN |
| 50 | Croatia | EASTERN EUROPE | NaN |
| 66 | Faroe Islands | WESTERN EUROPE | NaN |
| 78 | Gibraltar | WESTERN EUROPE | NaN |
| 101 | Italy | WESTERN EUROPE | NaN |
| 115 | Lebanon | NEAR EAST | NaN |
| 118 | Libya | NORTHERN AFRICA | NaN |
| 120 | Lithuania | BALTICS | NaN |
| 121 | Luxembourg | WESTERN EUROPE | NaN |
| 129 | Malta | WESTERN EUROPE | NaN |
| 137 | Moldova | C.W. OF IND. STATES | NaN |
| 138 | Monaco | WESTERN EUROPE | NaN |
| 141 | Morocco | NORTHERN AFRICA | NaN |
| 145 | Nepal | ASIA (EX. NEAR EAST) | NaN |
| 169 | Russia | C.W. OF IND. STATES | NaN |
| 171 | Saint Helena | SUB-SAHARAN AFRICA | NaN |
| 174 | St Pierre & Miquelon | NORTHERN AMERICA | NaN |
| 177 | San Marino | WESTERN EUROPE | NaN |
| 181 | Serbia | EASTERN EUROPE | NaN |
| 186 | Slovenia | EASTERN EUROPE | NaN |
| 200 | Tanzania | SUB-SAHARAN AFRICA | NaN |
country_df[pd.isnull(country_df['Climate'])]['Region'].value_counts()
| WESTERN EUROPE | 7 |
| SUB-SAHARAN AFRICA | 3 |
| EASTERN EUROPE | 3 |
| NORTHERN AMERICA | 2 |
| NORTHERN AFRICA | 2 |
| C.W. OF IND. STATES | 2 |
| NEAR EAST | 1 |
| BALTICS | 1 |
| ASIA (EX. NEAR EAST) | 1 |
| Name: Region, dtype: int64 |
# the Region value has annoying whitespaces that need to be stripped
country_df['Region'] = country_df['Region'].apply(lambda x: x.strip())
# climate zones in western europe
country_df[country_df['Region'] == 'WESTERN EUROPE']['Climate'].value_counts()
# climate zones in SUB-SAHARAN AFRICA
country_df[country_df['Region'] == 'SUB-SAHARAN AFRICA']['Climate'].value_counts()
# climate zones in EASTERN EUROPE
country_df[country_df['Region'] == 'EASTERN EUROPE']['Climate'].value_counts()
# climate zones in NORTHERN AMERICA
country_df[country_df['Region'] == 'NORTHERN AMERICA']['Climate'].value_counts()
# climate zones in NORTHERN AFRICA
country_df[country_df['Region'] == 'NORTHERN AFRICA']['Climate'].value_counts()
# climate zones in C.W. OF IND. STATES
country_df[country_df['Region'] == 'C.W. OF IND. STATES']['Climate'].value_counts()
# climate zones in NEAR EAST
country_df[country_df['Region'] == 'NEAR EAST']['Climate'].value_counts()
# climate zones in BALTICS
country_df[country_df['Region'] == 'BALTICS']['Climate'].value_counts()
# climate zones in ASIA (EX. NEAR EAST)
country_df[country_df['Region'] == 'ASIA (EX. NEAR EAST)']['Climate'].value_counts()
# we can either use the top value to fill missing climate data points
# or use a mean value:
country_df['Climate'] = country_df['Climate'].fillna(country_df.groupby('Region')['Climate'].transform('mean'))
# there are more missing values, e.g. literacy:
country_df[pd.isnull(country_df['Literacy (%)'])][['Country', 'Region', 'Literacy (%)']]
| Country | Region | Literacy (%) | |
|---|---|---|---|
| 25 | Bosnia & Herzegovina | EASTERN EUROPE | NaN |
| 66 | Faroe Islands | WESTERN EUROPE | NaN |
| 74 | Gaza Strip | NEAR EAST | NaN |
| 78 | Gibraltar | WESTERN EUROPE | NaN |
| 80 | Greenland | NORTHERN AMERICA | NaN |
| 85 | Guernsey | WESTERN EUROPE | NaN |
| 99 | Isle of Man | WESTERN EUROPE | NaN |
| 104 | Jersey | WESTERN EUROPE | NaN |
| 108 | Kiribati | OCEANIA | NaN |
| 123 | Macedonia | EASTERN EUROPE | NaN |
| 134 | Mayotte | SUB-SAHARAN AFRICA | NaN |
| 144 | Nauru | OCEANIA | NaN |
| 185 | Slovakia | EASTERN EUROPE | NaN |
| 187 | Solomon Islands | OCEANIA | NaN |
| 209 | Tuvalu | OCEANIA | NaN |
| 220 | Virgin Islands | LATIN AMER. & CARIB | NaN |
| 222 | West Bank | NEAR EAST | NaN |
| 223 | Western Sahara | NORTHERN AFRICA | NaN |
# here we can also fill with mean values:
country_df['Literacy (%)'] = country_df['Literacy (%)'].fillna(country_df.groupby('Region')['Literacy (%)'].transform('mean'))
# the remaining rows with missing values can be dropped for now
country_df = country_df.dropna(axis=0)
country_df.isnull().sum()
| Country | 0 |
| Region | 0 |
| Population | 0 |
| Area (sq. mi.) | 0 |
| Pop. Density (per sq. mi.) | 0 |
| Coastline (coast/area ratio) | 0 |
| Net migration | 0 |
| Infant mortality (per 1000 births) | 0 |
| GDP ($ per capita) | 0 |
| Literacy (%) | 0 |
| Phones (per 1000) | 0 |
| Arable (%) | 0 |
| Crops (%) | 0 |
| Other (%) | 0 |
| Climate | 0 |
| Birthrate | 0 |
| Deathrate | 0 |
| Agriculture | 0 |
| Industry | 0 |
| Service | 0 |
| dtype: int64 |
# drop the country column as it is a unique
# classifier that will not help with clustering
country_df_dropped = country_df.drop(['Country'], axis=1)
# the region column is useful but needs to be encoded
country_df_dropped = pd.get_dummies(country_df_dropped)
country_df_dropped.head(5).transpose()
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| Population | 31056997.00 | 3581655.000 | 3.293009e+07 | 57794.00 | 71201.00 |
| Area (sq. mi.) | 647500.00 | 28748.000 | 2.381740e+06 | 199.00 | 468.00 |
| Pop. Density (per sq. mi.) | 48.00 | 124.600 | 1.380000e+01 | 290.40 | 152.10 |
| Coastline (coast/area ratio) | 0.00 | 1.260 | 4.000000e-02 | 58.29 | 0.00 |
| Net migration | 23.06 | -4.930 | -3.900000e-01 | -20.71 | 6.60 |
| Infant mortality (per 1000 births) | 163.07 | 21.520 | 3.100000e+01 | 9.27 | 4.05 |
| GDP ($ per capita) | 700.00 | 4500.000 | 6.000000e+03 | 8000.00 | 19000.00 |
| Literacy (%) | 36.00 | 86.500 | 7.000000e+01 | 97.00 | 100.00 |
| Phones (per 1000) | 3.20 | 71.200 | 7.810000e+01 | 259.50 | 497.20 |
| Arable (%) | 12.13 | 21.090 | 3.220000e+00 | 10.00 | 2.22 |
| Crops (%) | 0.22 | 4.420 | 2.500000e-01 | 15.00 | 0.00 |
| Other (%) | 87.65 | 74.490 | 9.653000e+01 | 75.00 | 97.78 |
| Climate | 1.00 | 3.000 | 1.000000e+00 | 2.00 | 3.00 |
| Birthrate | 46.60 | 15.110 | 1.714000e+01 | 22.46 | 8.71 |
| Deathrate | 20.34 | 5.220 | 4.610000e+00 | 3.27 | 6.25 |
| Agriculture | 0.38 | 0.232 | 1.010000e-01 | 0.00 | 0.00 |
| Industry | 0.24 | 0.188 | 6.000000e-01 | 0.00 | 0.00 |
| Service | 0.38 | 0.579 | 2.980000e-01 | 0.00 | 0.00 |
| Region_ASIA (EX. NEAR EAST) | 1.00 | 0.000 | 0.000000e+00 | 0.00 | 0.00 |
| Region_BALTICS | 0.00 | 0.000 | 0.000000e+00 | 0.00 | 0.00 |
| Region_C.W. OF IND. STATES | 0.00 | 0.000 | 0.000000e+00 | 0.00 | 0.00 |
| Region_EASTERN EUROPE | 0.00 | 1.000 | 0.000000e+00 | 0.00 | 0.00 |
| Region_LATIN AMER. & CARIB | 0.00 | 0.000 | 0.000000e+00 | 0.00 | 0.00 |
| Region_NEAR EAST | 0.00 | 0.000 | 0.000000e+00 | 0.00 | 0.00 |
| Region_NORTHERN AFRICA | 0.00 | 0.000 | 1.000000e+00 | 0.00 | 0.00 |
| Region_NORTHERN AMERICA | 0.00 | 0.000 | 0.000000e+00 | 0.00 | 0.00 |
| Region_OCEANIA | 0.00 | 0.000 | 0.000000e+00 | 1.00 | 0.00 |
| Region_SUB-SAHARAN AFRICA | 0.00 | 0.000 | 0.000000e+00 | 0.00 | 0.00 |
| Region_WESTERN EUROPE | 0.00 | 0.000 | 0.000000e+00 | 0.00 | 1.00 |
# to be able to compare all datapoints they need to be normalized
country_scaler = StandardScaler()
country_df_scaled = country_scaler.fit_transform(country_df_dropped)
Model Training
# finding a good k-value for number of cluster
ssd_country = []
for k in range(2,30):
model = KMeans(n_clusters=k, n_init='auto')
model.fit(country_df_scaled)
ssd_country.append(model.inertia_)
plt.figure(figsize=(10,5))
plt.title('SSD as a Function of Number of Cluster')
plt.plot(range(2,30), ssd_country, 'o--')
plt.savefig('assets/Scikit_Learn_83.webp', bbox_inches='tight')
plt.figure(figsize=(10,5))
plt.title('Difference in SSD as a Function of Number of Clusters')
pd.Series(ssd_country).diff().plot(kind='bar')
plt.savefig('assets/Scikit_Learn_84.webp', bbox_inches='tight')
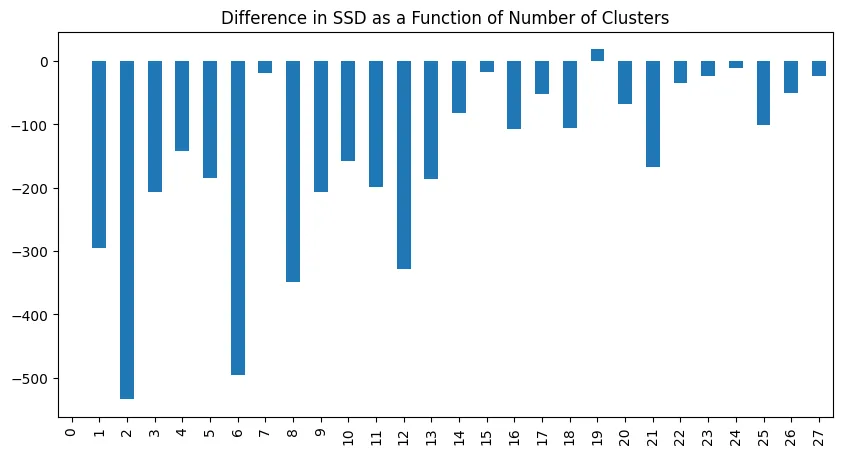
country_model = KMeans(
n_clusters=14,
n_init='auto',
random_state=42
)
# fit to find cluster centers and predict what center every feature belongs to
country_cluster_labels = country_model.fit_predict(country_df_scaled)
Model Evaluation
# add predicted label to source dataframe
country_df['Cluster14'] = country_cluster_labels
country_df['Cluster14'].value_counts()
plt.figure(figsize=(10, 7))
sns.set(style='darkgrid')
# hue/style by categorical column
sns.scatterplot(
x='GDP ($ per capita)',
y='Literacy (%)',
data=country_df,
s=40,
alpha=0.6,
hue='Cluster14',
palette='cool',
style='Region'
).set_title('Country Clusters with k=14')
plt.savefig('assets/Scikit_Learn_85.webp', bbox_inches='tight')
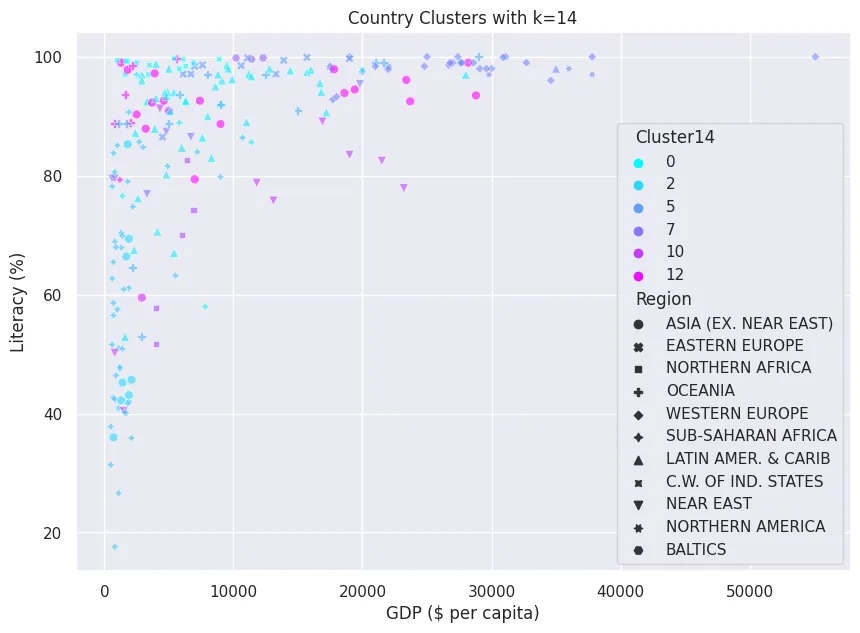
# repeat but only with 3 cluster
country_model2 = KMeans(
n_clusters=3,
n_init='auto',
random_state=42
)
# fit to find cluster centers and predict what center every feature belongs to
country_cluster_labels2 = country_model2.fit_predict(country_df_scaled)
# add predicted label to source dataframe
country_df['Cluster3'] = country_cluster_labels2
plt.figure(figsize=(10, 7))
sns.set(style='darkgrid')
# hue/style by categorical column
sns.scatterplot(
x='GDP ($ per capita)',
y='Literacy (%)',
data=country_df,
s=40,
alpha=0.6,
hue='Cluster3',
palette='cool',
style='Region'
).set_title('Country Clusters with k=3')
plt.savefig('assets/Scikit_Learn_86.webp', bbox_inches='tight')
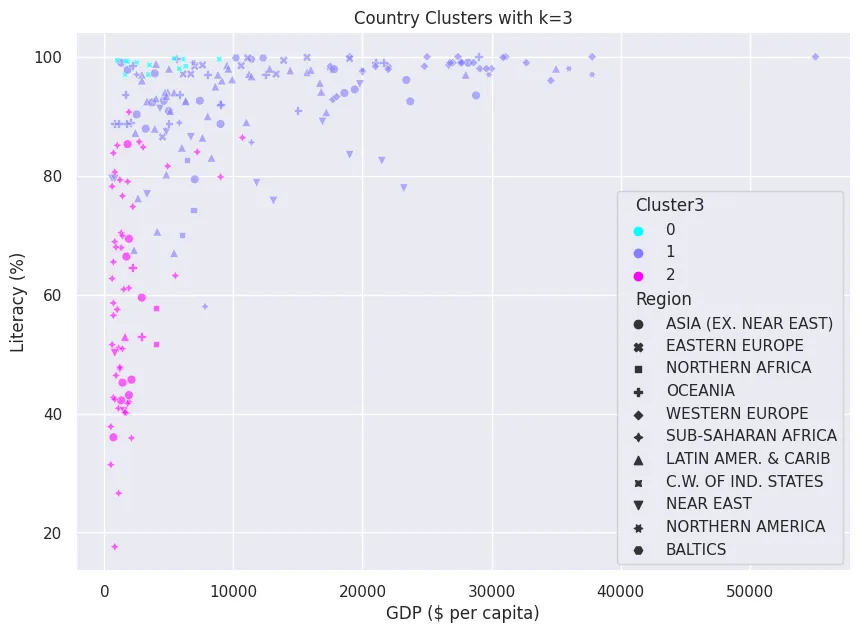
# How do the feature correlate with the predicted labels
country_label_corr = country_df.corr()['Cluster3']
print(country_label_corr.iloc[:-1].sort_values())
Feature Correlation
| Literacy (%) | -0.413704 |
| Crops (%) | -0.152936 |
| Coastline (coast/area ratio) | -0.132610 |
| Service | -0.070495 |
| Area (sq. mi.) | -0.062183 |
| Phones (per 1000) | -0.037538 |
| Population | -0.024969 |
| Industry | 0.008487 |
| Arable (%) | 0.034891 |
| Climate | 0.049659 |
| Other (%) | 0.050444 |
| Pop. Density (per sq. mi.) | 0.101062 |
| GDP ($ per capita) | 0.122206 |
| Agriculture | 0.250750 |
| Net migration | 0.316226 |
| Birthrate | 0.369940 |
| Infant mortality (per 1000 births) | 0.412365 |
| Deathrate | 0.575814 |
| Name: Cluster, dtype: float64 |
plt.figure(figsize=(10,6))
country_label_corr.iloc[:-1].sort_values().plot(kind='barh')
plt.title('Feature Importance')
plt.savefig('assets/Scikit_Learn_87.webp', bbox_inches='tight')
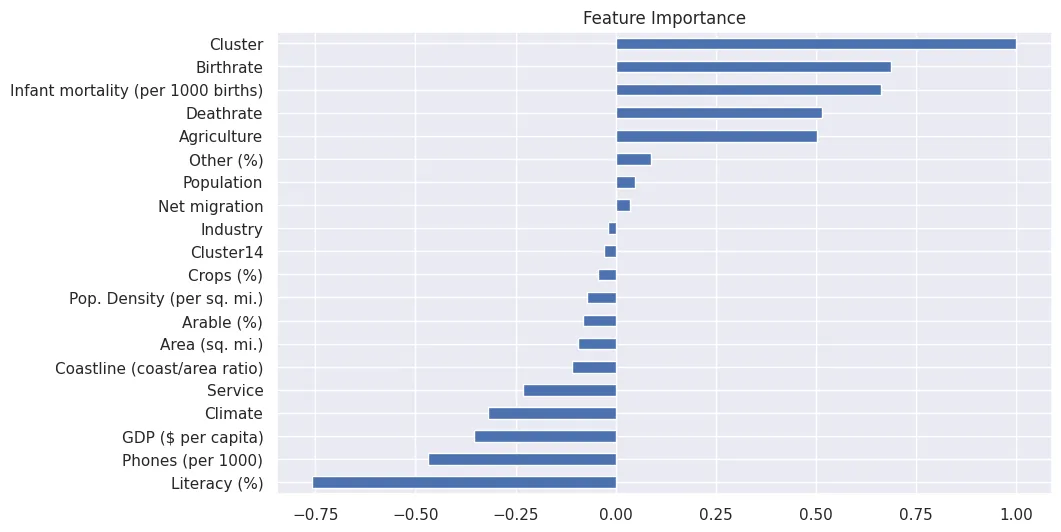
Plotly Choropleth Map
iso_codes = pd.read_csv('datasets/country-iso-codes.csv')
iso_map = iso_codes.set_index('Country')['ISO Code'].to_dict()
country_df['ISO Code'] = country_df['Country'].map(iso_map)
country_df[['Country','ISO Code']].head(5)
| Country | ISO Code | |
|---|---|---|
| 0 | Afghanistan | AFG |
| 1 | Albania | ALB |
| 2 | Algeria | DZA |
| 3 | American Samoa | ASM |
| 4 | Andorra | AND |
fig = px.choropleth(
country_df,
locations='ISO Code',
color='Cluster3',
hover_name='Country',
color_continuous_scale=px.colors.sequential.Plasma
)
fig.show()
fig = px.choropleth(
country_df,
locations='ISO Code',
color='Cluster14',
hover_name='Country',
color_continuous_scale=px.colors.sequential.Plasma
)
fig.show()
Unsupervised Learning - Agglomerative Clustering
Dataset Preprocessing
autompg_data: The Auto-MPG dataset for regression Revised from CMU StatLib library, data concerns city-cycle fuel consumption
autoMPG_df = pd.read_csv('datasets/auto-mpg.csv')
autoMPG_df.head(5)
| mpg | cylinders | displacement | horsepower | weight | acceleration | model_year | origin | name | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 18.0 | 8 | 307.0 | 130.0 | 3504 | 12.0 | 70 | usa | chevrolet chevelle malibu |
| 1 | 15.0 | 8 | 350.0 | 165.0 | 3693 | 11.5 | 70 | usa | buick skylark 320 |
| 2 | 18.0 | 8 | 318.0 | 150.0 | 3436 | 11.0 | 70 | usa | plymouth satellite |
| 3 | 16.0 | 8 | 304.0 | 150.0 | 3433 | 12.0 | 70 | usa | amc rebel sst |
| 4 | 17.0 | 8 | 302.0 | 140.0 | 3449 | 10.5 | 70 | usa | ford torino |
autoMPG_df['origin'].value_counts()
# there are only 3 countries of origin - can be turned into a dummy variable
autoMPG_dummy_df = pd.get_dummies(autoMPG_df.drop('name', axis=1))
autoMPG_dummy_df.head(5)
| mpg | cylinders | displacement | horsepower | weight | acceleration | model_year | origin_europe | origin_japan | origin_usa | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 18.0 | 8 | 307.0 | 130.0 | 3504 | 12.0 | 70 | False | False | True |
| 1 | 15.0 | 8 | 350.0 | 165.0 | 3693 | 11.5 | 70 | False | False | True |
| 2 | 18.0 | 8 | 318.0 | 150.0 | 3436 | 11.0 | 70 | False | False | True |
| 3 | 16.0 | 8 | 304.0 | 150.0 | 3433 | 12.0 | 70 | False | False | True |
| 4 | 17.0 | 8 | 302.0 | 140.0 | 3449 | 10.5 | 70 | False | False | True |
# normalize dataset
scaler = MinMaxScaler()
autoMPG_scaled = pd.DataFrame(
scaler.fit_transform(autoMPG_dummy_df), columns=autoMPG_dummy_df.columns
)
autoMPG_scaled.describe()
| mpg | cylinders | displacement | horsepower | weight | acceleration | model_year | origin_europe | origin_japan | origin_usa | |
|---|---|---|---|---|---|---|---|---|---|---|
| count | 392.000000 | 392.000000 | 392.000000 | 392.000000 | 392.000000 | 392.000000 | 392.000000 | 392.000000 | 392.000000 | 392.000000 |
| mean | 0.384200 | 0.494388 | 0.326646 | 0.317768 | 0.386897 | 0.448888 | 0.498299 | 0.173469 | 0.201531 | 0.625000 |
| std | 0.207580 | 0.341157 | 0.270398 | 0.209191 | 0.240829 | 0.164218 | 0.306978 | 0.379136 | 0.401656 | 0.484742 |
| min | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 0.212766 | 0.200000 | 0.095607 | 0.157609 | 0.173589 | 0.343750 | 0.250000 | 0.000000 | 0.000000 | 0.000000 |
| 50% | 0.365691 | 0.200000 | 0.214470 | 0.258152 | 0.337539 | 0.446429 | 0.500000 | 0.000000 | 0.000000 | 1.000000 |
| 75% | 0.531915 | 1.000000 | 0.536822 | 0.434783 | 0.567550 | 0.537202 | 0.750000 | 0.000000 | 0.000000 | 1.000000 |
| max | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
plt.figure(figsize=(12,10))
sns.heatmap(autoMPG_scaled, annot=False, cmap='viridis')
plt.savefig('assets/Scikit_Learn_90.webp', bbox_inches='tight')
sns.clustermap(
autoMPG_scaled.corr(numeric_only=True),
linewidth=0.5,
cmap='seismic',
annot=True,
col_cluster=False
)
plt.savefig('assets/Scikit_Learn_91.webp', bbox_inches='tight')
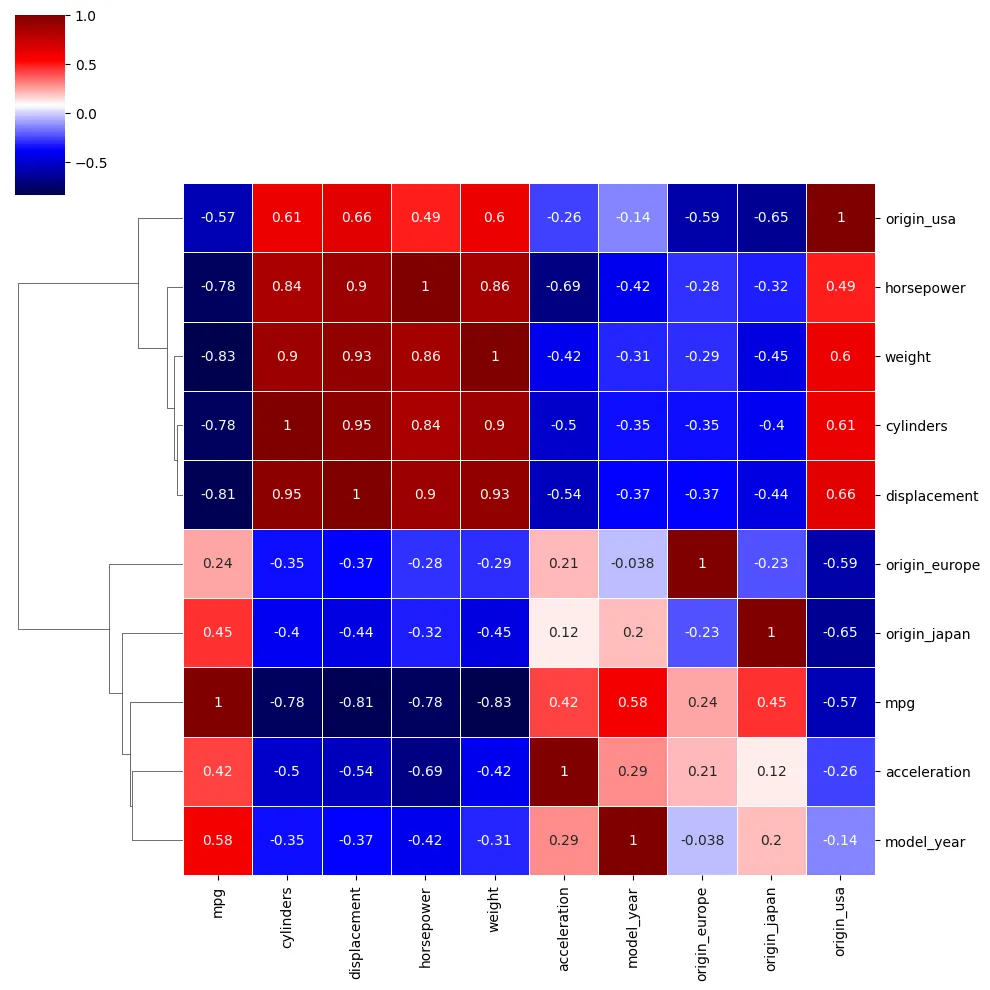
Assigning Cluster Labels
Known Number of Clusters
# there are ~ 4 clusters visible - let's try to agglomerate them
autoMPG_model = AgglomerativeClustering(n_clusters=4)
cluster_labels = autoMPG_model.fit_predict(autoMPG_scaled)
autoMPG_df['label'] = cluster_labels
autoMPG_df.head(5)
| mpg | cylinders | displacement | horsepower | weight | acceleration | model_year | origin | name | label | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 18.0 | 8 | 307.0 | 130.0 | 3504 | 12.0 | 70 | usa | chevrolet chevelle malibu | 2 |
| 1 | 15.0 | 8 | 350.0 | 165.0 | 3693 | 11.5 | 70 | usa | buick skylark 320 | 2 |
| 2 | 18.0 | 8 | 318.0 | 150.0 | 3436 | 11.0 | 70 | usa | plymouth satellite | 2 |
| 3 | 16.0 | 8 | 304.0 | 150.0 | 3433 | 12.0 | 70 | usa | amc rebel sst | 2 |
| 4 | 17.0 | 8 | 302.0 | 140.0 | 3449 | 10.5 | 70 | usa | ford torino | 2 |
plt.figure(figsize=(12,5))
sns.scatterplot(
x='mpg',
y='horsepower',
data=autoMPG_df,
hue='label',
palette='cool_r',
style='origin'
).set_title('Horsepower as a function of Miles-per-gallon')
plt.savefig('assets/Scikit_Learn_92.webp', bbox_inches='tight')
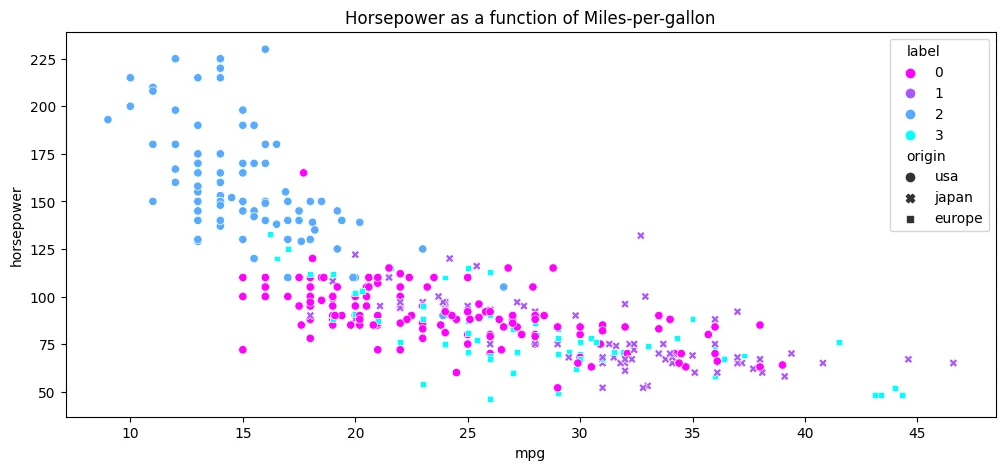
plt.figure(figsize=(12,5))
sns.scatterplot(
x='model_year',
y='mpg',
data=autoMPG_df,
hue='label',
palette='cool_r',
style='origin'
).set_title('Model Year as a function of Miles-per-gallon')
plt.legend(bbox_to_anchor=(1.01,1.01))
plt.savefig('assets/Scikit_Learn_93.webp', bbox_inches='tight')
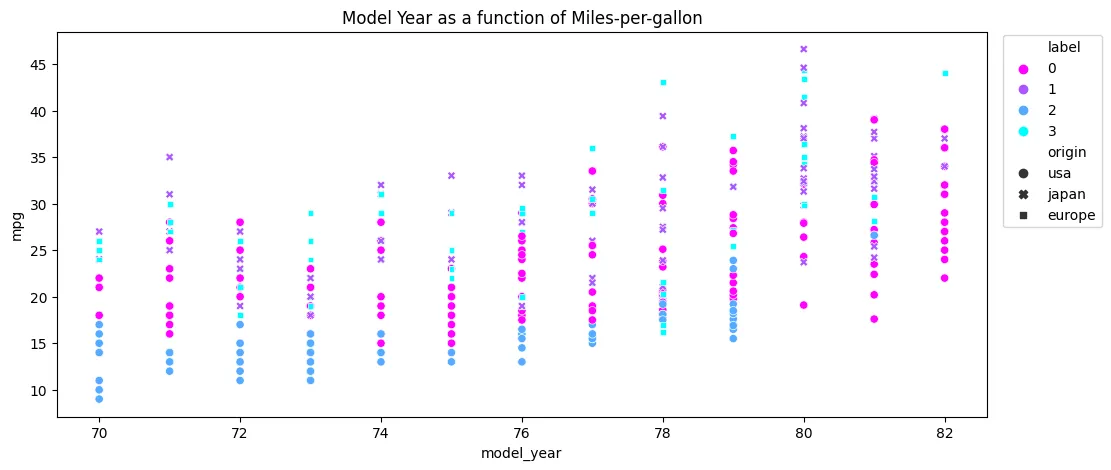
figure, axes = plt.subplots(1, 3, sharex=True,figsize=(15, 5))
figure.suptitle('Country of Origin')
axes[0].set_title('second chart with no data')
sns.scatterplot(
x='horsepower',
y='mpg',
data=autoMPG_df[autoMPG_df['origin'] == 'europe'],
hue='label',
palette='cool_r',
style='model_year',
ax=axes[0]
).set_title('Europe')
axes[1].set_title('Europe')
sns.scatterplot(
x='horsepower',
y='mpg',
data=autoMPG_df[autoMPG_df['origin'] == 'japan'],
hue='label',
palette='cool_r',
style='model_year',
ax=axes[1]
).set_title('Japan')
axes[2].set_title('second chart with no data')
sns.scatterplot(
x='horsepower',
y='mpg',
data=autoMPG_df[autoMPG_df['origin'] == 'usa'],
hue='label',
palette='cool_r',
style='model_year',
ax=axes[2]
).set_title('USA')
plt.legend(bbox_to_anchor=(1.01,1.01))
plt.savefig('assets/Scikit_Learn_94.webp', bbox_inches='tight')
# nice... perfect separation by country!
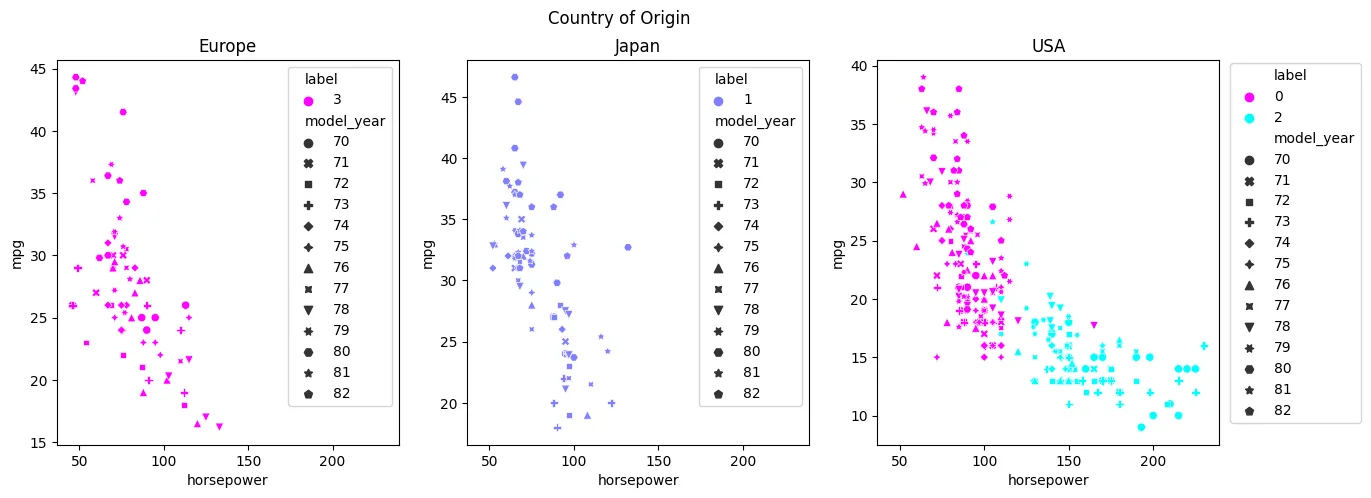
Unknown Number of Clusters
The Clustermap created above allowed us to estimate the amount of clusters needed to accuratly label the dataset based on the Dendrogram displayed on the left side. If we do not know how many clusters are present in our dataset we can define a maximum distance threshold a cluster can have before being merged with surrounding clusters. Setting this threshold to zero results in a number of clusters == number of datapoints.
autoMPG_model_auto = AgglomerativeClustering(
n_clusters=None,
metric='euclidean',
distance_threshold=0
)
cluster_labels_auto = autoMPG_model_auto.fit_predict(autoMPG_scaled)
len(np.unique(cluster_labels_auto))
# threshold of zero leads to 392 clusters == number of rows in our dataset
# find out a good distance threshold
linkage_matrix = hierarchy.linkage(autoMPG_model_auto.children_)
linkage_matrix
# [`cluster[i]`, `cluster[j]`, `distance between`, `number of members`]
# to display this matrix we can use the above mentioned dendrogram
plt.figure(figsize=(20,10))
plt.title('Hierarchy Dendrogram for 8 Classes')
dendro = hierarchy.dendrogram(linkage_matrix, truncate_mode='lastp', p=9)
plt.savefig('assets/Scikit_Learn_95.webp', bbox_inches='tight')
# The higher the y-value the larger the distance between the connected clusters
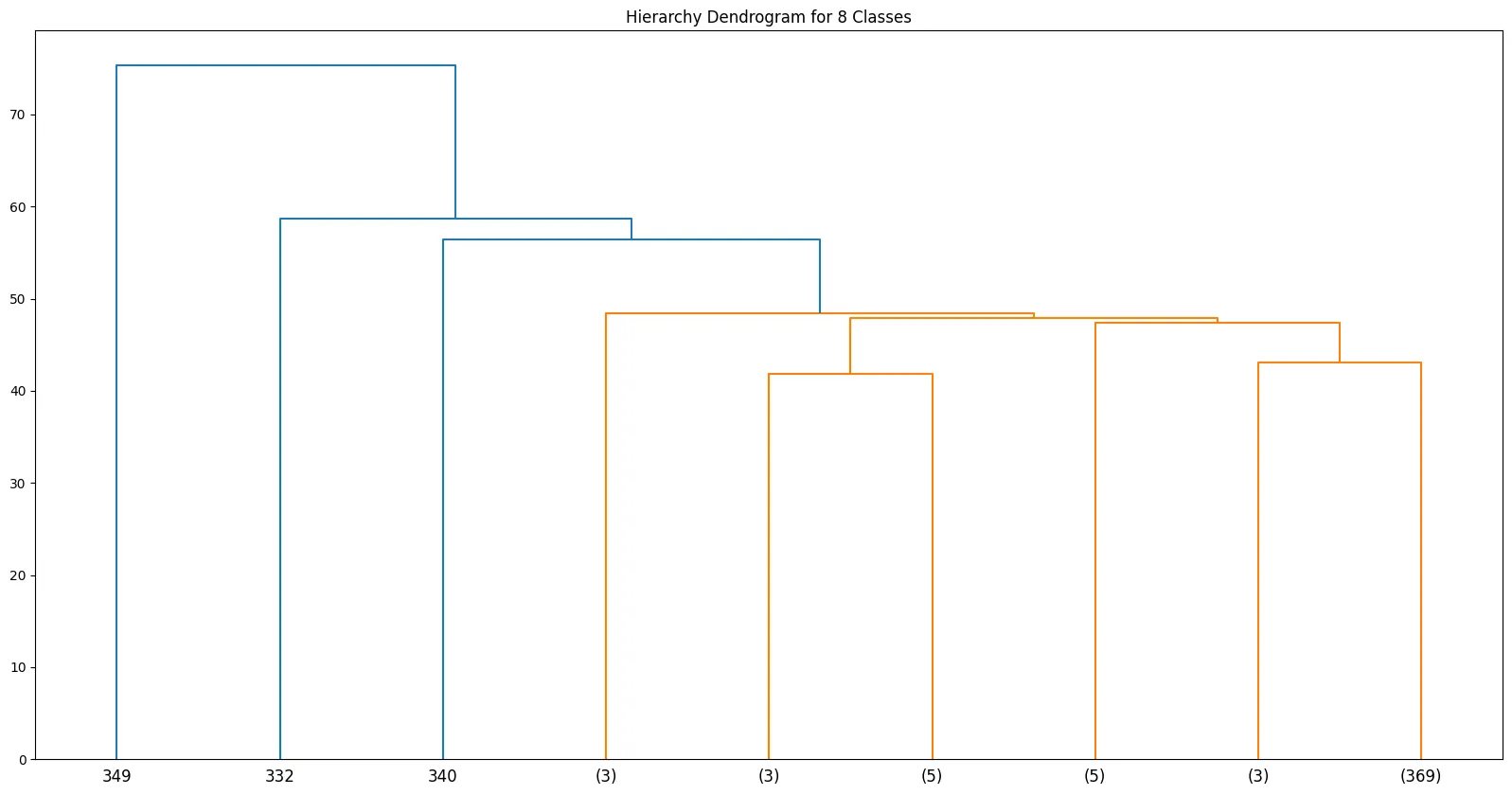
# since the miles-per-gallons are a good indicator for the label
# what is the max distance between two points here:
car_max_mpg = autoMPG_scaled.iloc[autoMPG_scaled['mpg'].idxmax()]
car_min_mpg = autoMPG_scaled.iloc[autoMPG_scaled['mpg'].idxmin()]
np.linalg.norm(car_max_mpg - car_min_mpg)
# 3.1128158766165406
# if the max distance is ~3 the threshold should be < 3
autoMPG_model_auto = AgglomerativeClustering(
n_clusters=None,
metric='euclidean',
distance_threshold=2
)
cluster_labels_auto = autoMPG_model_auto.fit_predict(autoMPG_scaled)
len(np.unique(cluster_labels_auto))
# threshold of two leads to 11 clusters
autoMPG_model_auto = AgglomerativeClustering(
n_clusters=None,
metric='euclidean',
distance_threshold=3
)
cluster_labels_auto = autoMPG_model_auto.fit_predict(autoMPG_scaled)
len(np.unique(cluster_labels_auto))
# threshold of three leads to 9 clusters
autoMPG_df['label_auto'] = cluster_labels_auto
figure, axes = plt.subplots(1, 3, sharex=True,figsize=(15, 6))
figure.suptitle('Country of Origin')
axes[0].set_title('second chart with no data')
sns.scatterplot(
x='horsepower',
y='mpg',
data=autoMPG_df[autoMPG_df['origin'] == 'europe'],
hue='label_auto',
palette='cool_r',
style='model_year',
ax=axes[0]
).set_title('Europe')
axes[1].set_title('Europe')
sns.scatterplot(
x='horsepower',
y='mpg',
data=autoMPG_df[autoMPG_df['origin'] == 'japan'],
hue='label_auto',
palette='cool_r',
style='model_year',
ax=axes[1]
).set_title('Japan')
axes[2].set_title('second chart with no data')
sns.scatterplot(
x='horsepower',
y='mpg',
data=autoMPG_df[autoMPG_df['origin'] == 'usa'],
hue='label_auto',
palette='cool_r',
style='model_year',
ax=axes[2]
).set_title('USA')
plt.legend(bbox_to_anchor=(1.01,1.01))
plt.savefig('assets/Scikit_Learn_96.webp', bbox_inches='tight')
# the division by countries is still there. but we are now getting
# sub-classes within each country - which might be important depending on your set goal
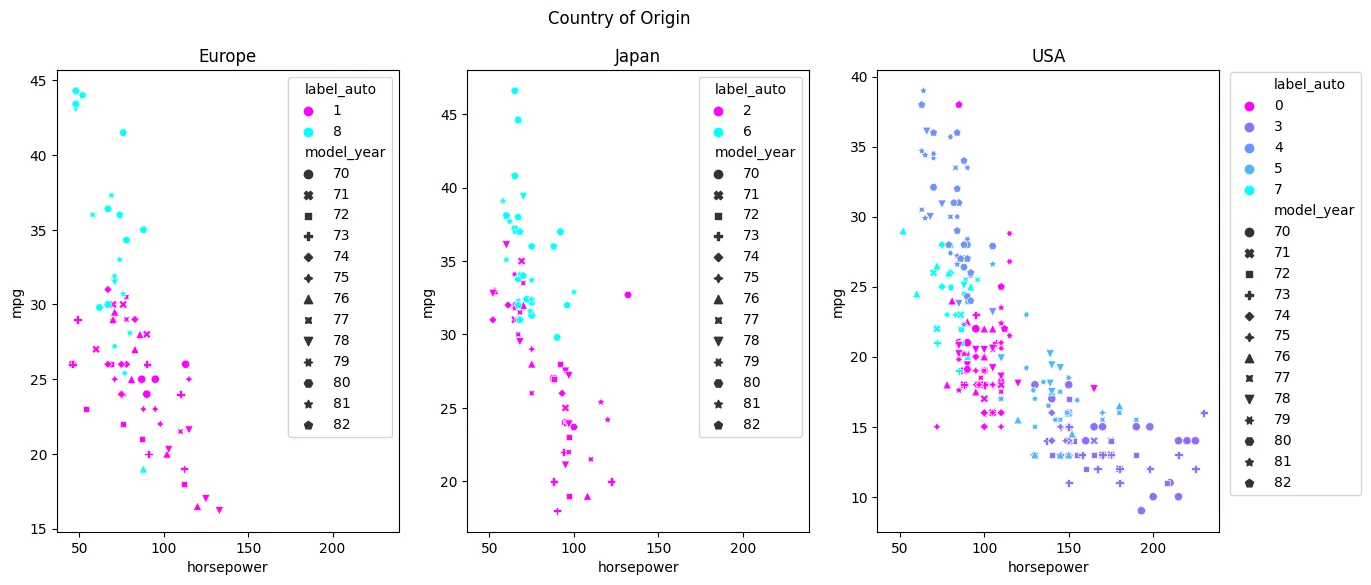
Unsupervised Learning - Density-based Spatial Clustering (DBSCAN)
DBSCAN vs KMeans
blobs_df = pd.read_csv('datasets/blobs.csv')
blobs_df.tail(2)
| X1 | X2 | |
|---|---|---|
| 1498 | 5.454552 | 6.461246 |
| 1499 | -7.769230 | 7.014384 |
plt.figure(figsize=(12,5))
plt.title('Blobs Dataset')
sns.scatterplot(data=blobs_df, x='X1', y='X2')
plt.savefig('assets/Scikit_Learn_97.webp', bbox_inches='tight')
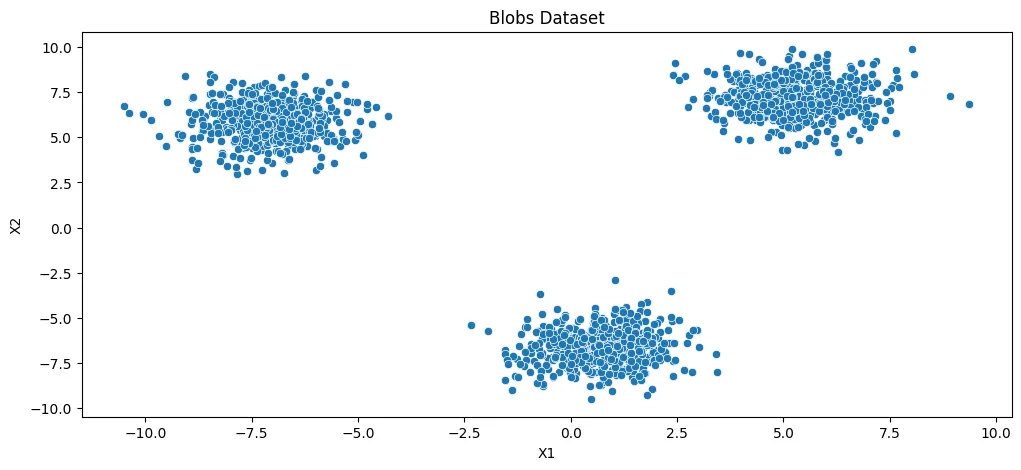
moons_df = pd.read_csv('datasets/moons.csv')
moons_df.tail(2)
| X1 | X2 | |
|---|---|---|
| 1498 | 1.803858 | -0.154705 |
| 1499 | 0.203305 | 0.079049 |
plt.figure(figsize=(12,5))
plt.title('Moons Dataset')
sns.scatterplot(data=moons_df, x='X1', y='X2')
plt.savefig('assets/Scikit_Learn_98.webp', bbox_inches='tight')
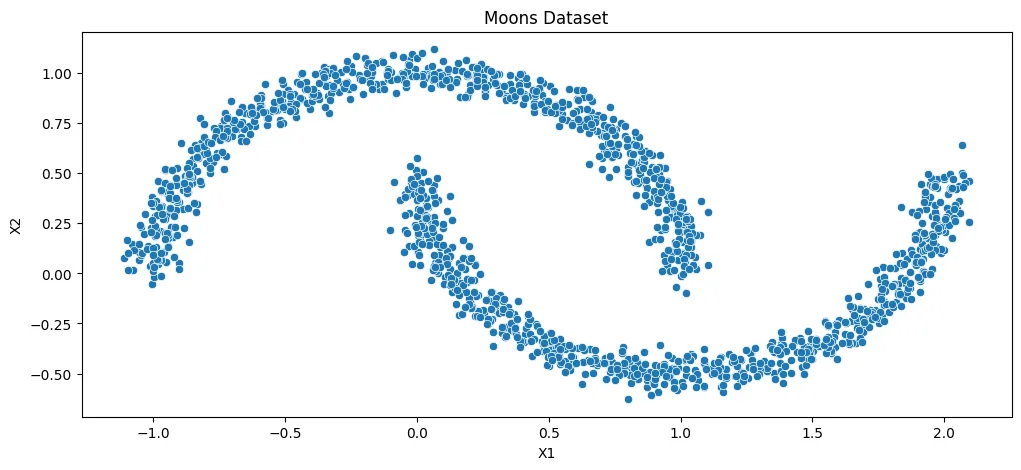
circles_df = pd.read_csv('datasets/circles.csv')
circles_df.tail(2)
| X1 | X2 | |
|---|---|---|
| 1498 | 0.027432 | -0.264891 |
| 1499 | -0.216732 | 0.183006 |
plt.figure(figsize=(12,5))
plt.title('Circles Dataset')
sns.scatterplot(data=circles_df, x='X1', y='X2')
plt.savefig('assets/Scikit_Learn_99.webp', bbox_inches='tight')
def display_categories(model, data, axis):
labels = model.fit_predict(data)
sns.scatterplot(data=data, x='X1', y='X2', hue=labels, palette='cool' , ax=axis)
km_model_blobs = KMeans(n_clusters=3, init='random', n_init='auto')
db_model_blobs = DBSCAN(eps=0.5, min_samples=5)
figure, axes = plt.subplots(1, 2, sharex=True,figsize=(12, 6))
figure.suptitle('3 Blobs Dataset')
axes[0].set_title('KMeans Clustering')
display_categories(km_model_blobs, blobs_df, axes[0])
axes[1].set_title('DBSCAN Clustering')
display_categories(db_model_blobs, blobs_df, axes[1])
plt.savefig('assets/Scikit_Learn_100.webp', bbox_inches='tight')
km_model_moons = KMeans(n_clusters=2, init='random', n_init='auto')
db_model_moons = DBSCAN(eps=0.2, min_samples=5)
figure, axes = plt.subplots(1, 2, sharex=True,figsize=(12, 6))
figure.suptitle('2 Moons Dataset')
axes[0].set_title('KMeans Clustering')
display_categories(km_model_moons, moons_df, axes[0])
axes[1].set_title('DBSCAN Clustering')
display_categories(db_model_moons, moons_df, axes[1])
plt.savefig('assets/Scikit_Learn_101.webp', bbox_inches='tight')
km_model_circles = KMeans(n_clusters=2, init='random', n_init='auto')
db_model_circles = DBSCAN(eps=0.2, min_samples=5)
figure, axes = plt.subplots(1, 2, sharex=True,figsize=(12, 6))
figure.suptitle('2 Circles Dataset')
axes[0].set_title('KMeans Clustering')
display_categories(km_model_circles, circles_df, axes[0])
axes[1].set_title('DBSCAN Clustering')
display_categories(db_model_circles, circles_df, axes[1])
plt.savefig('assets/Scikit_Learn_102.webp', bbox_inches='tight')
DBSCAN Hyperparameter Tuning
two_blobs_df = pd.read_csv('datasets/two-blobs.csv')
two_blobs_otl_df = pd.read_csv('datasets/two-blobs-outliers.csv')
# default hyperparameter
db_model_base = DBSCAN(eps=0.5, min_samples=5)
figure, axes = plt.subplots(1, 2, sharex=True,figsize=(12, 6))
figure.suptitle('2 Blobs Dataset - Default Hyperparameter')
axes[0].set_title('DBSCAN Clustering w/o Outliers')
display_categories(db_model_base, two_blobs_df, axes[0])
axes[1].set_title('DBSCAN Clustering with Outliers')
display_categories(db_model_base, two_blobs_otl_df, axes[1])
plt.savefig('assets/Scikit_Learn_103.webp', bbox_inches='tight')
# points around cluster 1 are assigned to be outliers
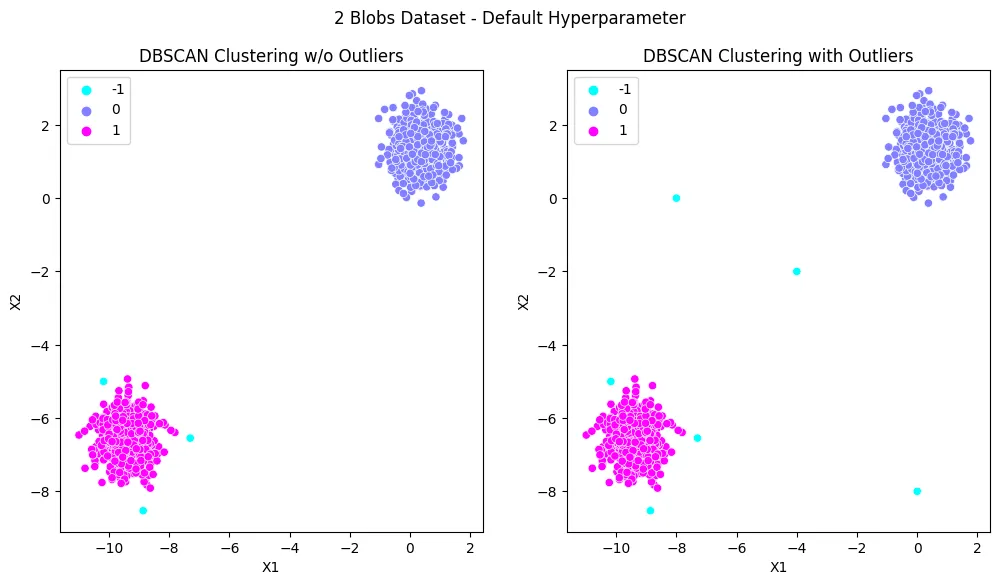
# reducing epsilon reduces the max distance (epsilon)
# points are allowed to have and still be assigned to a cluster
db_model_dec = DBSCAN(eps=0.001, min_samples=5)
figure, axes = plt.subplots(1, 2, sharex=True,figsize=(12, 6))
figure.suptitle('2 Blobs Dataset - Reduced Epsilon')
axes[0].set_title('DBSCAN Clustering w/o Outliers')
display_categories(db_model_dec, two_blobs_df, axes[0])
axes[1].set_title('DBSCAN Clustering with Outliers')
display_categories(db_model_dec, two_blobs_otl_df, axes[1])
plt.savefig('assets/Scikit_Learn_104.webp', bbox_inches='tight')
# distance is too small - every point becomes it's own cluster and is assigned as an outlier
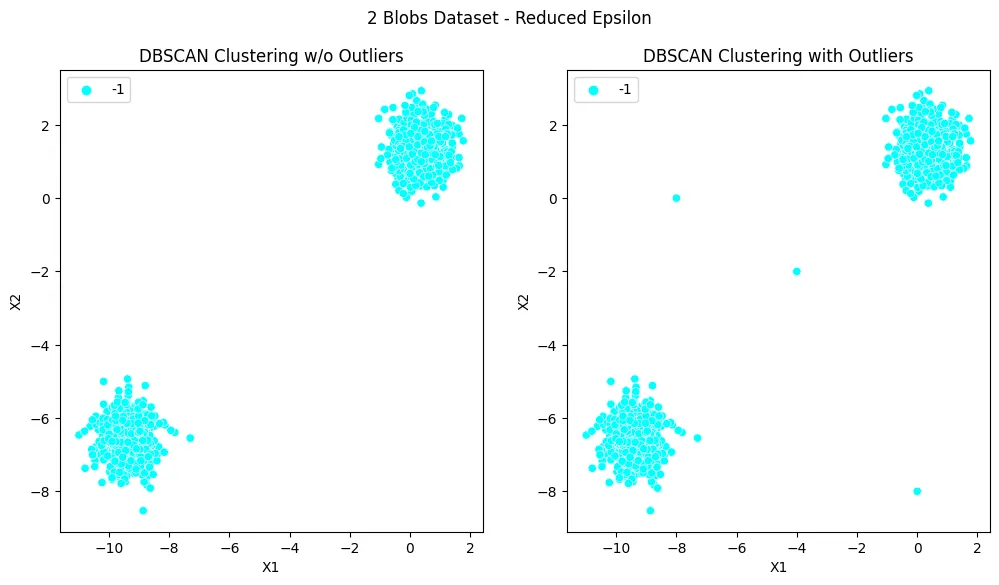
# increasing epsilon increases the max distance (epsilon)
# points are allowed to have and still be assigned to a cluster
db_model_inc = DBSCAN(eps=10, min_samples=5)
figure, axes = plt.subplots(1, 2, sharex=True,figsize=(12, 6))
figure.suptitle('2 Blobs Dataset - Increased Epsilon')
axes[0].set_title('DBSCAN Clustering w/o Outliers')
display_categories(db_model_inc, two_blobs_df, axes[0])
axes[1].set_title('DBSCAN Clustering with Outliers')
display_categories(db_model_inc, two_blobs_otl_df, axes[1])
plt.savefig('assets/Scikit_Learn_105.webp', bbox_inches='tight')
# distance is too big - every point becomes becomes part of the same cluster
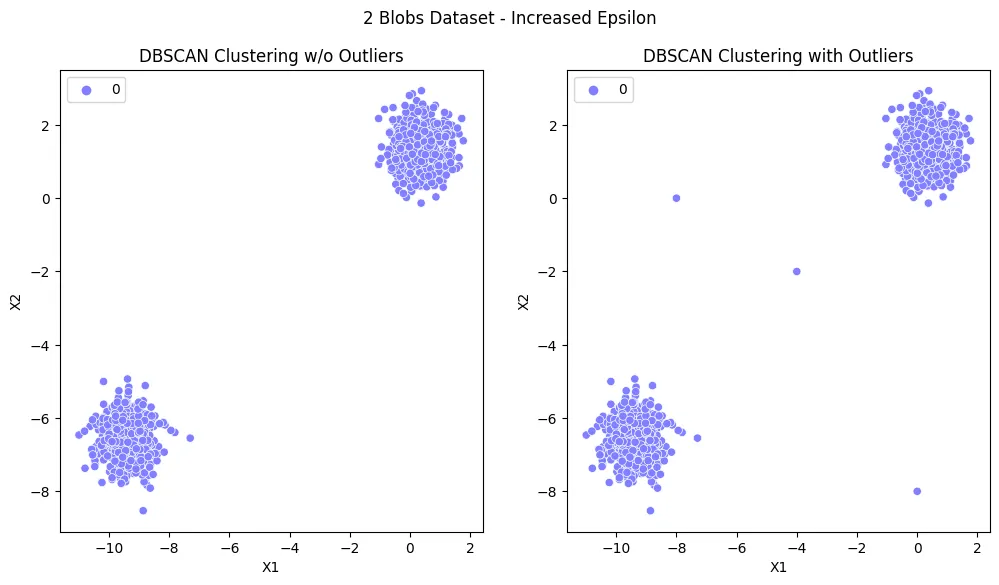
Elbow Plot
epsilon_value_range = np.linspace(0.0001, 1, 100)
n_outliers = []
perc_outlier = []
n_clusters = []
for epsilon in epsilon_value_range:
dbscan_model = DBSCAN(eps=epsilon)
dbscan_model.fit(two_blobs_otl_df)
# total number of outliers
n_outliers.append(np.sum(dbscan_model.labels_ == -1))
# percentage of outliers
perc_outlier.append(
100 * np.sum(dbscan_model.labels_ == -1) / len(dbscan_model.labels_)
)
# number of clusters
n_clusters.append(len(np.unique(dbscan_model.labels_)))
plt.figure(figsize=(12,5))
plt.title('Elbow Plot - DBSCAN Hyperparameter')
plt.xlabel('Epsilon (Max Distance between Points)')
plt.ylabel('Number of Outliers')
plt.ylim(0,10)
# we expect 3 outliers
plt.hlines(y=3, xmin=0, xmax=0.7, color='fuchsia')
# 3 outliers are reached somewhere around eps=0.7
plt.vlines(x=0.7, ymin=0, ymax=3, color='fuchsia')
sns.lineplot(x=epsilon_value_range, y=n_outliers)
plt.savefig('assets/Scikit_Learn_107.webp', bbox_inches='tight')
plt.figure(figsize=(12,5))
plt.title('Number of Clusters by Epsilon Range')
plt.xlabel('Epsilon (Max Distance between Points)')
plt.ylabel('Number of Clusters')
# we expect 2 clusters + outliers
plt.hlines(y=3, xmin=0, xmax=1, color='fuchsia')
plt.ylim(0,50)
plt.xlim(0,1)
sns.lineplot(x=epsilon_value_range, y=n_clusters)
plt.savefig('assets/Scikit_Learn_108.webp', bbox_inches='tight')
# we already reach 3 cluster with an epsilon of 0.2
# but as seen above we need an epsilon of 0.7 to reduce
# the number of outliers to 3
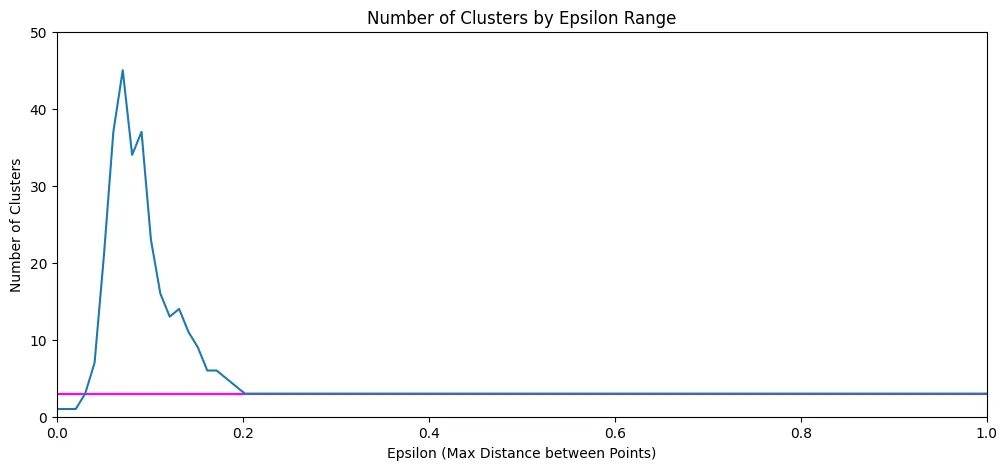
# find the optimum
# rule of thumb for min_samples = 2*n_dim
n_dim = two_blobs_otl_df.shape[1]
db_model_opt = DBSCAN(eps=0.7, min_samples=2*n_dim)
figure, axes = plt.subplots(1, 2, sharex=True,figsize=(12, 6))
figure.suptitle('2 Blobs Dataset - Optimal Epsilon')
axes[0].set_title('DBSCAN Clustering w/o Outliers')
display_categories(db_model_opt, two_blobs_df, axes[0])
axes[1].set_title('DBSCAN Clustering with Outliers')
display_categories(db_model_opt, two_blobs_otl_df, axes[1])
plt.savefig('assets/Scikit_Learn_106.webp', bbox_inches='tight')
# the 3 outliers are labled as such and every other point is assigned to one of the two clusters
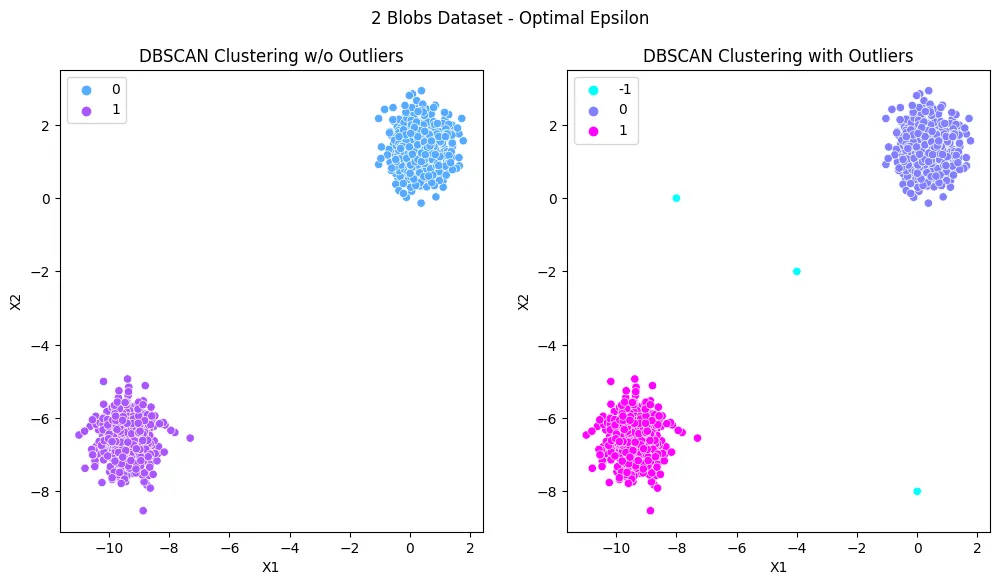
# find number of outliers
print('Number of Outliers', np.sum(db_model_opt.labels_ == -1))
# Number of Outliers 3
# get outlier percentage
print('Percentage of Outliers', (100 * np.sum(db_model_opt.labels_ == -1) / len(db_model_opt.labels_)).round(2),'%')
# Percentage of Outliers 0.3 %
Realworld Dataset
Wholesale customers The data set refers to clients of a wholesale distributor. It includes the annual spending in monetary units (m.u.) on diverse product categories
Additional Information
- FRESH: annual spending (m.u.) on fresh products (Continuous)
- MILK: annual spending (m.u.) on milk products (Continuous)
- GROCERY: annual spending (m.u.) on grocery products (Continuous)
- FROZEN: annual spending (m.u.) on frozen products (Continuous)
- DETERGENTS_PAPER: annual spending (m.u.) on detergents and paper products (Continuous)
- DELICATESSEN: annual spending (m.u.)on and delicatessen products (Continuous)
- CHANNEL: customers Channel - Horeca (Hotel/Restaurant/Cafe) or Retail channel (Nominal)
- REGION: customers Region - Lisnon, Oporto or Other (Nominal)
Dataset Exploration
wholesale_df = pd.read_csv('datasets/wholesome-customers-data.csv')
wholesale_df.head(5)
| Channel | Region | Fresh | Milk | Grocery | Frozen | Detergents_Paper | Delicassen | |
|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 3 | 12669 | 9656 | 7561 | 214 | 2674 | 1338 |
| 1 | 2 | 3 | 7057 | 9810 | 9568 | 1762 | 3293 | 1776 |
| 2 | 2 | 3 | 6353 | 8808 | 7684 | 2405 | 3516 | 7844 |
| 3 | 1 | 3 | 13265 | 1196 | 4221 | 6404 | 507 | 1788 |
| 4 | 2 | 3 | 22615 | 5410 | 7198 | 3915 | 1777 | 5185 |
wholesale_df.info()
plt.figure(figsize=(12,5))
plt.title('Whole Sale: Milk Products vs Groceries')
sns.scatterplot(
data=wholesale_df,
x='Milk', y='Grocery',
hue='Channel', style='Region',
palette='winter'
)
plt.savefig('assets/Scikit_Learn_109.webp', bbox_inches='tight')
plt.figure(figsize=(10, 5))
plt.title('Whole Sale: Milk Products by Distribution Channel')
sns.histplot(
data=wholesale_df,
x='Milk',
bins=50,
hue='Channel',
palette='winter',
kde=True
)
plt.savefig('assets/Scikit_Learn_110.webp', bbox_inches='tight')
sns.clustermap(
wholesale_df.corr(),
linewidth=0.5,
cmap='winter',
annot=True,
col_cluster=False
)
plt.savefig('assets/Scikit_Learn_111.webp', bbox_inches='tight')
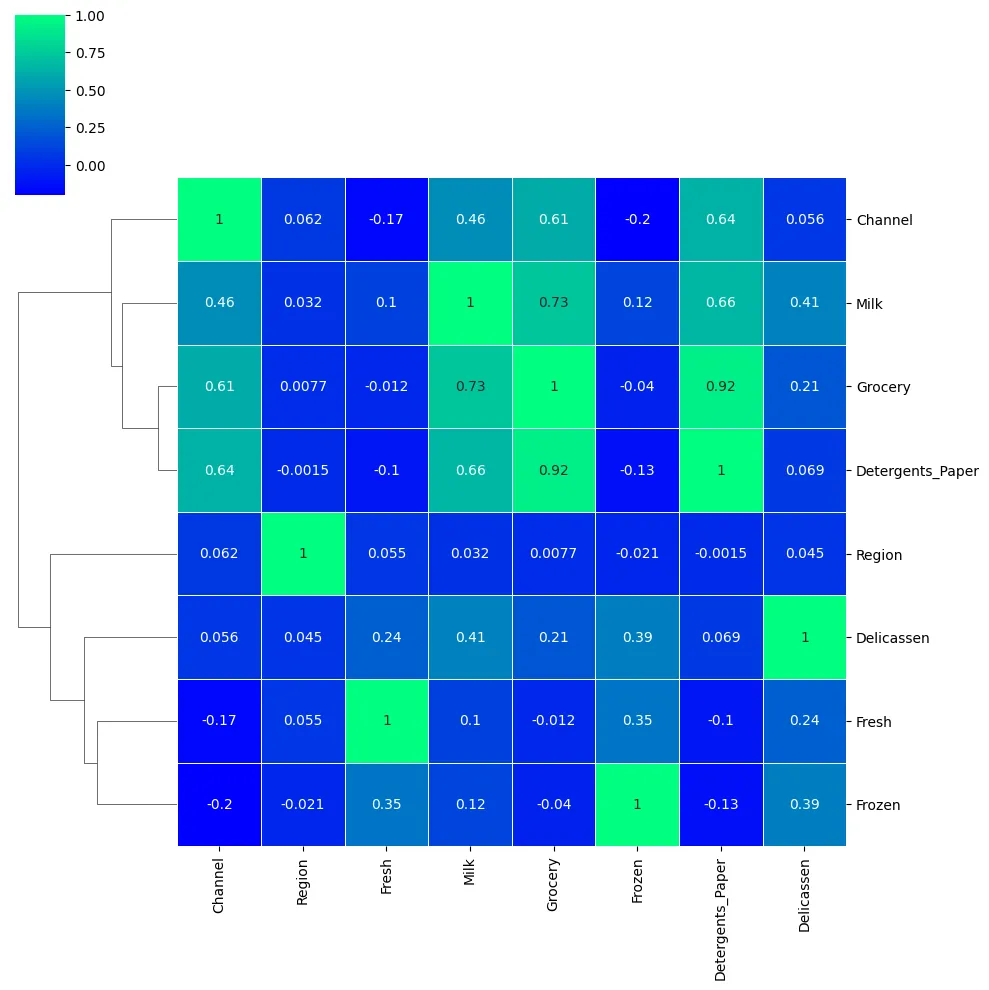
sns.pairplot(
data=wholesale_df,
hue='Region',
palette='winter'
)
plt.savefig('assets/Scikit_Learn_112.webp', bbox_inches='tight')
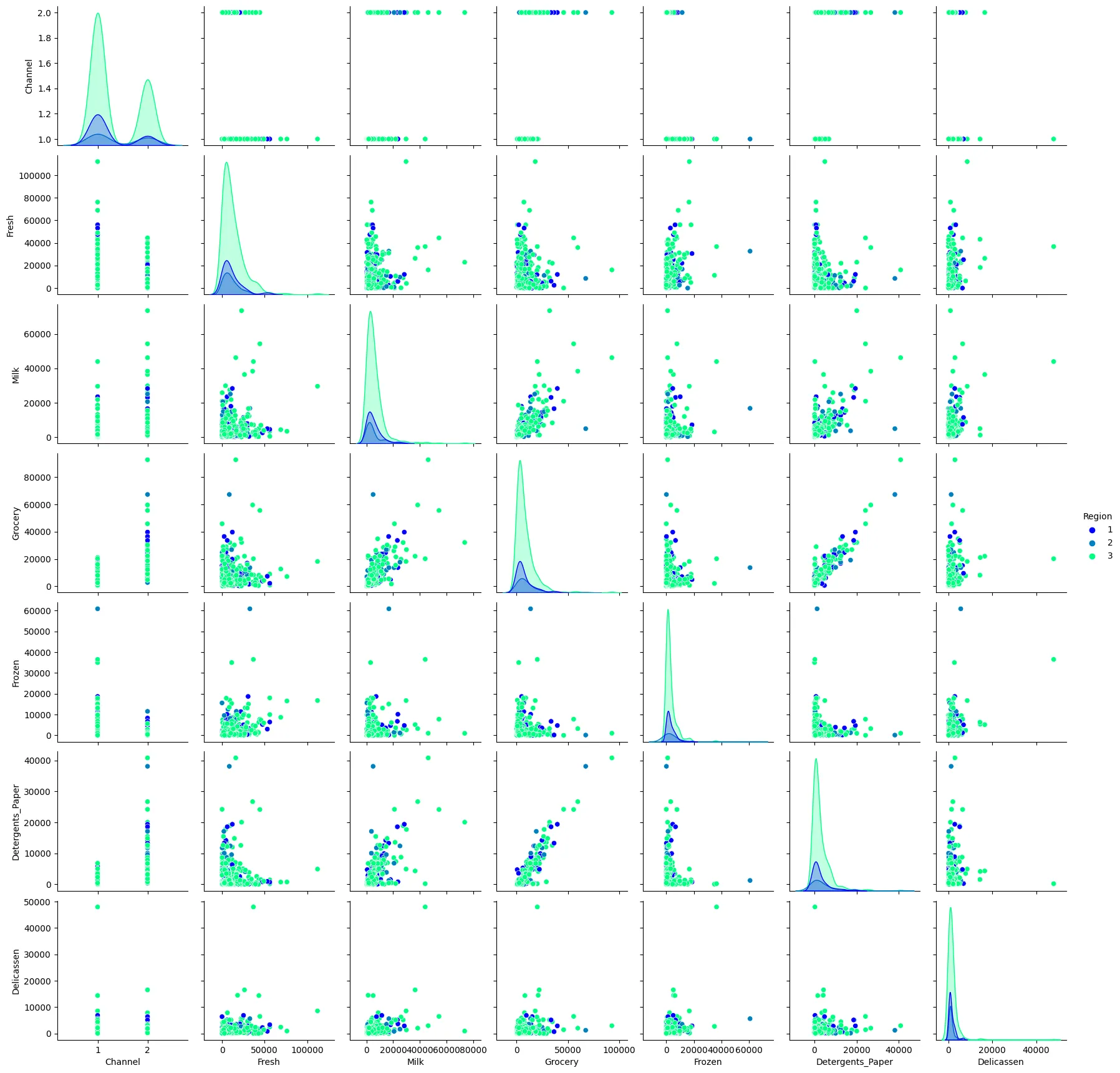
Data Preprocessing
# normalize feature set
scaler = StandardScaler()
wholesale_scaled = pd.DataFrame(
scaler.fit_transform(wholesale_df), columns=wholesale_df.columns
)
wholesale_scaled.describe()
| Channel | Region | Fresh | Milk | Grocery | Frozen | Detergents_Paper | Delicassen | |
|---|---|---|---|---|---|---|---|---|
| count | 4.400000e+02 | 4.400000e+02 | 4.400000e+02 | 440.000000 | 4.400000e+02 | 4.400000e+02 | 4.400000e+02 | 4.400000e+02 |
| mean | 1.614870e-17 | 3.552714e-16 | -3.431598e-17 | 0.000000 | -4.037175e-17 | 3.633457e-17 | 2.422305e-17 | -8.074349e-18 |
| std | 1.001138e+00 | 1.001138e+00 | 1.001138e+00 | 1.001138 | 1.001138e+00 | 1.001138e+00 | 1.001138e+00 | 1.001138e+00 |
| min | -6.902971e-01 | -1.995342e+00 | -9.496831e-01 | -0.778795 | -8.373344e-01 | -6.283430e-01 | -6.044165e-01 | -5.402644e-01 |
| 25% | -6.902971e-01 | -7.023369e-01 | -7.023339e-01 | -0.578306 | -6.108364e-01 | -4.804306e-01 | -5.511349e-01 | -3.964005e-01 |
| 50% | -6.902971e-01 | 5.906683e-01 | -2.767602e-01 | -0.294258 | -3.366684e-01 | -3.188045e-01 | -4.336004e-01 | -1.985766e-01 |
| 75% | 1.448652e+00 | 5.906683e-01 | 3.905226e-01 | 0.189092 | 2.849105e-01 | 9.946441e-02 | 2.184822e-01 | 1.048598e-01 |
| max | 1.448652e+00 | 5.906683e-01 | 7.927738e+00 | 9.183650 | 8.936528e+00 | 1.191900e+01 | 7.967672e+00 | 1.647845e+01 |
Model Hyperparameter Tuning
epsilon_value_range = np.linspace(0.001, 3, 100)
n_dim = wholesale_scaled.shape[1]
n_outliers = []
perc_outlier = []
n_clusters = []
for epsilon in epsilon_value_range:
dbscan_model = DBSCAN(eps=epsilon, min_samples=2*n_dim)
dbscan_model.fit(wholesale_scaled)
# total number of outliers
n_outliers.append(np.sum(dbscan_model.labels_ == -1))
# percentage of outliers
perc_outlier.append(
100 * np.sum(dbscan_model.labels_ == -1) / len(dbscan_model.labels_)
)
# number of clusters
n_clusters.append(len(np.unique(dbscan_model.labels_)))
plt.figure(figsize=(12,5))
plt.title('Elbow Plot - DBSCAN Hyperparameter')
plt.xlabel('Epsilon (Max Distance between Points)')
plt.ylabel('Number of Outliers')
plt.hlines(y=25, xmin=0, xmax=2, color='fuchsia')
plt.vlines(x=2, ymin=0, ymax=25, color='fuchsia')
sns.lineplot(x=epsilon_value_range, y=n_outliers)
plt.savefig('assets/Scikit_Learn_113.webp', bbox_inches='tight')
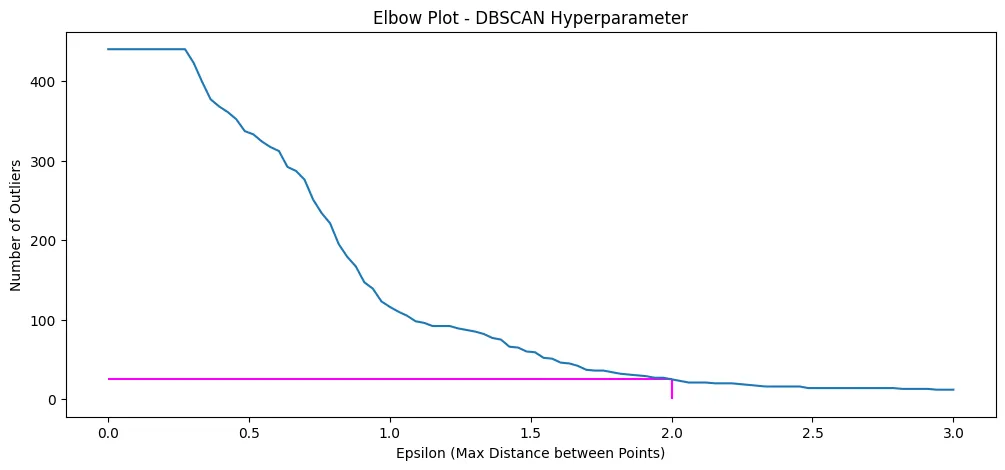
plt.figure(figsize=(12,5))
plt.title('Number of Clusters by Epsilon Range')
plt.xlabel('Epsilon (Max Distance between Points)')
plt.ylabel('Number of Clusters')
plt.hlines(y=3, xmin=0, xmax=2, color='fuchsia')
plt.vlines(x=2, ymin=0, ymax=3, color='fuchsia')
sns.lineplot(x=epsilon_value_range, y=n_clusters)
plt.savefig('assets/Scikit_Learn_114.webp', bbox_inches='tight')
def wholesale_categories(model, data, x, y, axis):
labels = model.fit_predict(data)
sns.scatterplot(data=data, x=x, y=y, hue=labels, palette='cool' , ax=axis)
db_model_opt = DBSCAN(eps=2.0, min_samples=2*n_dim)
figure, axes = plt.subplots(1, 2, sharex=True,figsize=(12, 6))
figure.suptitle('Whole Sale Dataset - DBSCAN Cluster (Normalized)')
axes[0].set_title('DBSCAN Clustering Milk Products vs Groceries')
wholesale_categories(
model=db_model_opt,
data=wholesale_scaled,
x='Milk', y='Grocery',
axis=axes[0]
)
axes[1].set_title('DBSCAN Clustering Milk Products vs Delicassen')
wholesale_categories(
model=db_model_opt,
data=wholesale_scaled,
x='Milk', y='Delicassen',
axis=axes[1]
)
plt.savefig('assets/Scikit_Learn_115a.webp', bbox_inches='tight')
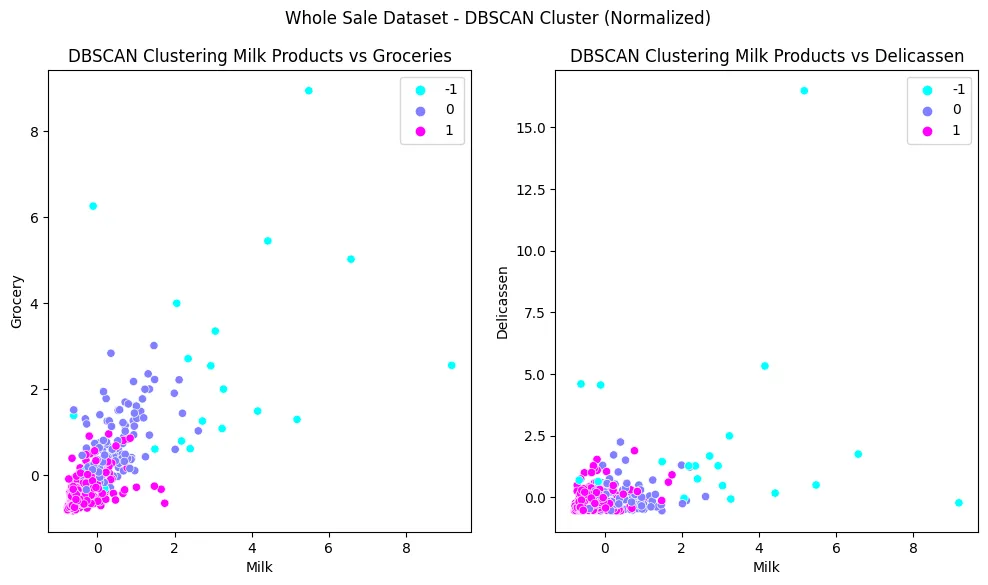
# add labels to original dataframe
wholesale_df['Label'] = db_model_opt.fit_predict(wholesale_scaled)
wholesale_df['Label'].head(5)
# remove outliers
wholesale_df_wo_otl = wholesale_df[wholesale_df['Label'] != -1]
db_model_opt = DBSCAN(eps=3.0, min_samples=2*n_dim)
figure, axes = plt.subplots(1, 2, sharex=True,figsize=(12, 6))
figure.suptitle('Whole Sale Dataset - DBSCAN Cluster (w/o Outliers)')
axes[0].set_title('DBSCAN Clustering Milk Products vs Groceries')
sns.scatterplot(
data=wholesale_df_wo_otl,
x='Milk', y='Grocery',
hue='Label',
palette='cool',
ax=axes[0]
)
axes[1].set_title('DBSCAN Clustering Milk Products vs Delicassen')
sns.scatterplot(
data=wholesale_df_wo_otl,
x='Milk', y='Delicassen',
hue='Label',
palette='cool',
ax=axes[1]
)
plt.savefig('assets/Scikit_Learn_115b.webp', bbox_inches='tight')
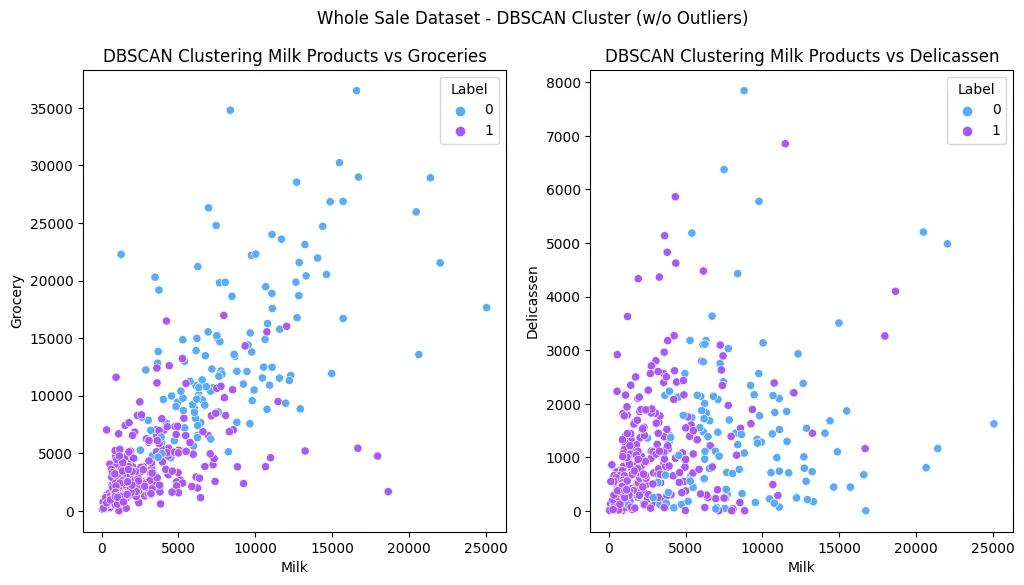
# see if the mean values of each cluster differ from each other
grouped_df = wholesale_df.groupby('Label').mean()
| Label | Channel | Region | Fresh | Milk | Grocery | Frozen | Detergents_Paper | Delicassen |
|---|---|---|---|---|---|---|---|---|
| -1 | 1.52 | 2.480000 | 27729.920000 | 22966.960000 | 26609.600000 | 11289.640000 | 11173.560000 | 6707.160000 |
| 0 | 2.00 | 2.620155 | 8227.666667 | 8615.852713 | 13859.674419 | 1447.759690 | 5969.581395 | 1498.457364 |
| 1 | 1.00 | 2.513986 | 12326.972028 | 3023.559441 | 3655.328671 | 3086.181818 | 763.783217 | 1083.786713 |
scaler = MinMaxScaler()
grouped_scaler = pd.DataFrame(
scaler.fit_transform(grouped_df), columns=grouped_df.columns, index=['Outlier', 'Cluster 1', 'Cluster 2']
)
grouped_scaler.head()
| Channel | Region | Fresh | Milk | Grocery | Frozen | Detergents_Paper | Delicassen | |
|---|---|---|---|---|---|---|---|---|
| Outlier | 0.52 | 0.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
| Cluster 1 | 1.00 | 1.000000 | 0.000000 | 0.280408 | 0.444551 | 0.000000 | 0.500087 | 0.073741 |
| Cluster 2 | 0.00 | 0.242489 | 0.210196 | 0.000000 | 0.000000 | 0.166475 | 0.000000 | 0.000000 |
plt.figure(figsize=(12, 3))
plt.title('Scaled Cluster / Outliers Comparison (Normalized)')
sns.heatmap(
grouped_scaler,
linewidth=0.5,
cmap='coolwarm',
annot=True
)
plt.savefig('assets/Scikit_Learn_116.webp', bbox_inches='tight')
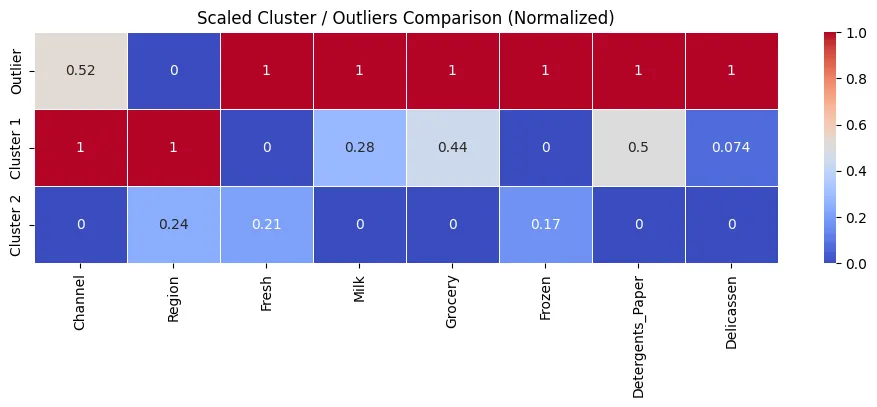
grouped_df = grouped_df.drop(['Labels'], axis=1)
# remove outlier
wholesale_clusters = grouped_df.drop(-1, axis=0)
wholesale_clusters.head()
| Label | Channel | Region | Fresh | Milk | Grocery | Frozen | Detergents_Paper | Delicassen |
|---|---|---|---|---|---|---|---|---|
| 0 | 2.0 | 2.620155 | 8227.666667 | 8615.852713 | 13859.674419 | 1447.759690 | 5969.581395 | 1498.457364 |
| 1 | 1.0 | 2.513986 | 12326.972028 | 3023.559441 | 3655.328671 | 3086.181818 | 763.783217 | 1083.786713 |
plt.figure(figsize=(12, 3))
plt.title('Mean Spending Values for Cluster 1 and 2')
sns.heatmap(
wholesale_clusters,
linewidth=0.5,
cmap='coolwarm',
annot=True
)
plt.savefig('assets/Scikit_Learn_117.webp', bbox_inches='tight')
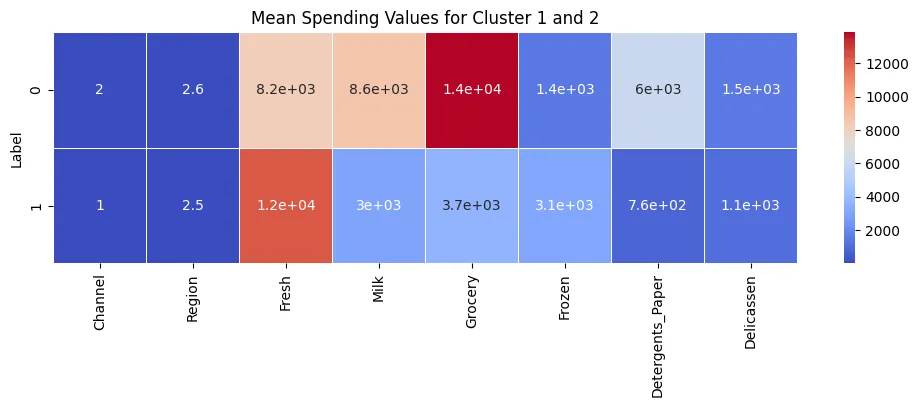
Dimensionality Reduction - Principal Component Analysis (PCA)
Dataset Preprocessing
Breast cancer wisconsin (diagnostic) dataset.
- Attribute Information:
- radius (mean of distances from center to points on the perimeter)
- texture (standard deviation of gray-scale values)
- perimeter
- area
- smoothness (local variation in radius lengths)
- compactness (perimeter^2 / area - 1.0)
- concavity (severity of concave portions of the contour)
- concave points (number of concave portions of the contour)
- symmetry
- fractal dimension ("coastline approximation" - 1)
The mean, standard error, and "worst" or largest (mean of the three worst/largest values) of these features were computed for each image, resulting in 30 features. For instance, field 0 is Mean Radius, field 10 is Radius SE, field 20 is Worst Radius.
- class:
- WDBC-Malignant
- WDBC-Benign
tumor_df = pd.read_csv('datasets/cancer-tumor-data-features.csv')
tumor_df.head(5).transpose()
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| mean radius | 17.990000 | 20.570000 | 19.690000 | 11.420000 | 20.290000 |
| mean texture | 10.380000 | 17.770000 | 21.250000 | 20.380000 | 14.340000 |
| mean perimeter | 122.800000 | 132.900000 | 130.000000 | 77.580000 | 135.100000 |
| mean area | 1001.000000 | 1326.000000 | 1203.000000 | 386.100000 | 1297.000000 |
| mean smoothness | 0.118400 | 0.084740 | 0.109600 | 0.142500 | 0.100300 |
| mean compactness | 0.277600 | 0.078640 | 0.159900 | 0.283900 | 0.132800 |
| mean concavity | 0.300100 | 0.086900 | 0.197400 | 0.241400 | 0.198000 |
| mean concave points | 0.147100 | 0.070170 | 0.127900 | 0.105200 | 0.104300 |
| mean symmetry | 0.241900 | 0.181200 | 0.206900 | 0.259700 | 0.180900 |
| mean fractal dimension | 0.078710 | 0.056670 | 0.059990 | 0.097440 | 0.058830 |
| radius error | 1.095000 | 0.543500 | 0.745600 | 0.495600 | 0.757200 |
| texture error | 0.905300 | 0.733900 | 0.786900 | 1.156000 | 0.781300 |
| perimeter error | 8.589000 | 3.398000 | 4.585000 | 3.445000 | 5.438000 |
| area error | 153.400000 | 74.080000 | 94.030000 | 27.230000 | 94.440000 |
| smoothness error | 0.006399 | 0.005225 | 0.006150 | 0.009110 | 0.011490 |
| compactness error | 0.049040 | 0.013080 | 0.040060 | 0.074580 | 0.024610 |
| concavity error | 0.053730 | 0.018600 | 0.038320 | 0.056610 | 0.056880 |
| concave points error | 0.015870 | 0.013400 | 0.020580 | 0.018670 | 0.018850 |
| symmetry error | 0.030030 | 0.013890 | 0.022500 | 0.059630 | 0.017560 |
| fractal dimension error | 0.006193 | 0.003532 | 0.004571 | 0.009208 | 0.005115 |
| worst radius | 25.380000 | 24.990000 | 23.570000 | 14.910000 | 22.540000 |
| worst texture | 17.330000 | 23.410000 | 25.530000 | 26.500000 | 16.670000 |
| worst perimeter | 184.600000 | 158.800000 | 152.500000 | 98.870000 | 152.200000 |
| worst area | 2019.000000 | 1956.000000 | 1709.000000 | 567.700000 | 1575.000000 |
| worst smoothness | 0.162200 | 0.123800 | 0.144400 | 0.209800 | 0.137400 |
| worst compactness | 0.665600 | 0.186600 | 0.424500 | 0.866300 | 0.205000 |
| worst concavity | 0.711900 | 0.241600 | 0.450400 | 0.686900 | 0.400000 |
| worst concave points | 0.265400 | 0.186000 | 0.243000 | 0.257500 | 0.162500 |
| worst symmetry | 0.460100 | 0.275000 | 0.361300 | 0.663800 | 0.236400 |
| worst fractal dimension | 0.118900 | 0.089020 | 0.087580 | 0.173000 | 0.076780 |
# normalizing data
scaler = StandardScaler()
tumor_scaled_arr = scaler.fit_transform(tumor_df)
tumor_scaled_df = pd.DataFrame(
tumor_scaled_arr, columns=tumor_df.columns
)
tumor_scaled_df.head(5).transpose()
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| mean radius | 1.097064 | 1.829821 | 1.579888 | -0.768909 | 1.750297 |
| mean texture | -2.073335 | -0.353632 | 0.456187 | 0.253732 | -1.151816 |
| mean perimeter | 1.269934 | 1.685955 | 1.566503 | -0.592687 | 1.776573 |
| mean area | 0.984375 | 1.908708 | 1.558884 | -0.764464 | 1.826229 |
| mean smoothness | 1.568466 | -0.826962 | 0.942210 | 3.283553 | 0.280372 |
| mean compactness | 3.283515 | -0.487072 | 1.052926 | 3.402909 | 0.539340 |
| mean concavity | 2.652874 | -0.023846 | 1.363478 | 1.915897 | 1.371011 |
| mean concave points | 2.532475 | 0.548144 | 2.037231 | 1.451707 | 1.428493 |
| mean symmetry | 2.217515 | 0.001392 | 0.939685 | 2.867383 | -0.009560 |
| mean fractal dimension | 2.255747 | -0.868652 | -0.398008 | 4.910919 | -0.562450 |
| radius error | 2.489734 | 0.499255 | 1.228676 | 0.326373 | 1.270543 |
| texture error | -0.565265 | -0.876244 | -0.780083 | -0.110409 | -0.790244 |
| perimeter error | 2.833031 | 0.263327 | 0.850928 | 0.286593 | 1.273189 |
| area error | 2.487578 | 0.742402 | 1.181336 | -0.288378 | 1.190357 |
| smoothness error | -0.214002 | -0.605351 | -0.297005 | 0.689702 | 1.483067 |
| compactness error | 1.316862 | -0.692926 | 0.814974 | 2.744280 | -0.048520 |
| concavity error | 0.724026 | -0.440780 | 0.213076 | 0.819518 | 0.828471 |
| concave points error | 0.660820 | 0.260162 | 1.424827 | 1.115007 | 1.144205 |
| symmetry error | 1.148757 | -0.805450 | 0.237036 | 4.732680 | -0.361092 |
| fractal dimension error | 0.907083 | -0.099444 | 0.293559 | 2.047511 | 0.499328 |
| worst radius | 1.886690 | 1.805927 | 1.511870 | -0.281464 | 1.298575 |
| worst texture | -1.359293 | -0.369203 | -0.023974 | 0.133984 | -1.466770 |
| worst perimeter | 2.303601 | 1.535126 | 1.347475 | -0.249939 | 1.338539 |
| worst area | 2.001237 | 1.890489 | 1.456285 | -0.550021 | 1.220724 |
| worst smoothness | 1.307686 | -0.375612 | 0.527407 | 3.394275 | 0.220556 |
| worst compactness | 2.616665 | -0.430444 | 1.082932 | 3.893397 | -0.313395 |
| worst concavity | 2.109526 | -0.146749 | 0.854974 | 1.989588 | 0.613179 |
| worst concave points | 2.296076 | 1.087084 | 1.955000 | 2.175786 | 0.729259 |
| worst symmetry | 2.750622 | -0.243890 | 1.152255 | 6.046041 | -0.868353 |
| worst fractal dimension | 1.937015 | 0.281190 | 0.201391 | 4.935010 | -0.397100 |
Model Fitting
pca_model = PCA(n_components=2)
pca_results = pca_model.fit_transform(tumor_scaled_df)
print(pca_model.explained_variance_ratio_)
print(np.sum(pca_model.explained_variance_ratio_))
# the two principal components are able to describe
# 63% of the variance in the dataset
# [0.44272026 0.18971182]
# 0.6324320765155945
# adding components to original dataframe
tumor_df[['PC1','PC2']] = pca_results
tumor_df[['PC1','PC2']].head(5).transpose()
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| PC1 | 9.192837 | 2.387802 | 5.733896 | 7.122953 | 3.935302 |
| PC2 | 1.948583 | -3.768172 | -1.075174 | 10.275589 | -1.948072 |
plt.figure(figsize=(12,5))
plt.title('Principal Component Analysis - Cancer Tumor Dataset')
sns.scatterplot(
data=tumor_df,
x='PC1', y='PC2'
)
plt.savefig('assets/Scikit_Learn_118.webp', bbox_inches='tight')
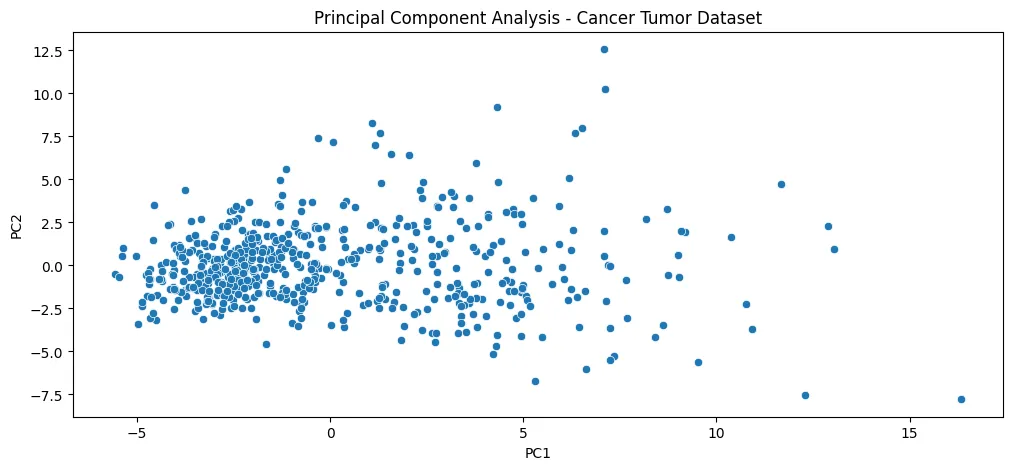
# get label data from dataset to confirm that we still have
# separably clusters after reducing the dimensions to 2
from sklearn.datasets import load_breast_cancer
tumor_dataset = load_breast_cancer()
tumor_dataset.keys()
# dict_keys(['data', 'target', 'frame', 'target_names', 'DESCR', 'feature_names', 'filename', 'data_module'])
tumor_dataset['target']
plt.figure(figsize=(12,5))
plt.title('PCA Cancer Tumor Dataset - Coloured by Labels')
sns.scatterplot(
data=tumor_df,
x='PC1', y='PC2',
hue=tumor_dataset['target'],
palette='winter'
)
plt.savefig('assets/Scikit_Learn_119.webp', bbox_inches='tight')
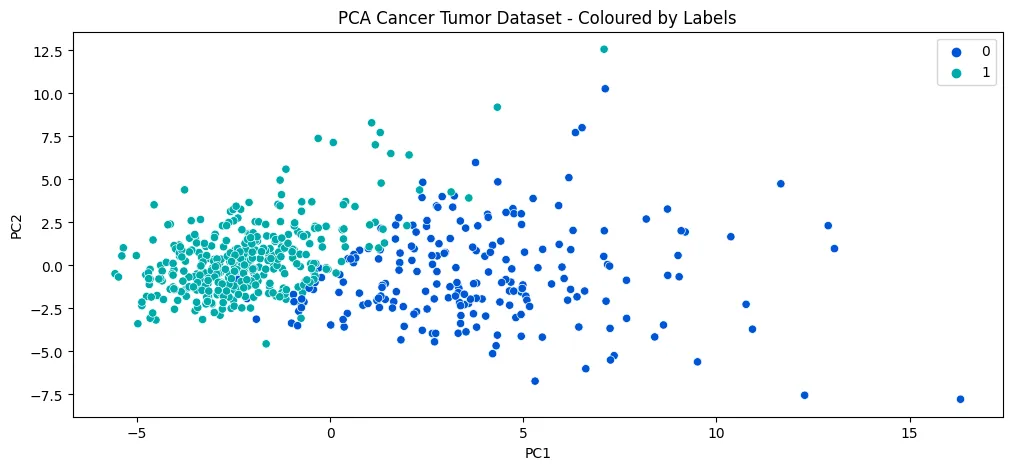
# as shown above we get around 63% of the variance explained by using 2 principal components
# since the dataset has 30 features 30 principal components will explain 100% of the variance
explained_variance = []
for n in range(1,31):
pca = PCA(n_components=n)
pca.fit(tumor_scaled_df)
explained_variance.append(np.sum(pca.explained_variance_ratio_))
plt.figure(figsize=(10, 5))
plt.title('Explained Variance by Number of Principal Components')
plt.xlabel('Principal Components')
sns.set(style='darkgrid')
sns.barplot(
data=pd.DataFrame(explained_variance, columns=['Explained Variance']),
x=np.arange(1,31),
y='Explained Variance'
)
plt.savefig('assets/Scikit_Learn_120.webp', bbox_inches='tight')
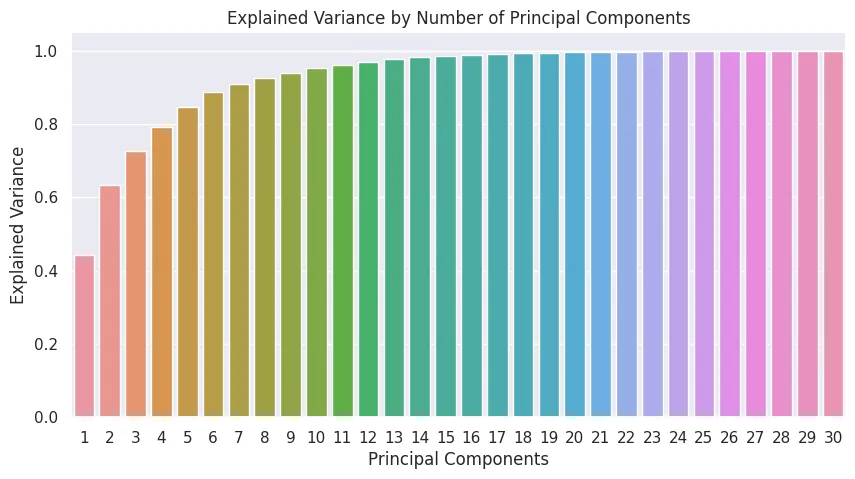
Dataset 2
What handwritten numbers are the hardest to tell apart for a ML Model?
digits_df = pd.read_csv('datasets/digits.csv')
digits_df.head(5).transpose()
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| pixel_0_0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| pixel_0_1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| pixel_0_2 | 5.0 | 0.0 | 0.0 | 7.0 | 0.0 |
| pixel_0_3 | 13.0 | 12.0 | 4.0 | 15.0 | 1.0 |
| pixel_0_4 | 9.0 | 13.0 | 15.0 | 13.0 | 11.0 |
| ... | |||||
| pixel_7_4 | 10.0 | 16.0 | 11.0 | 13.0 | 16.0 |
| pixel_7_5 | 0.0 | 10.0 | 16.0 | 9.0 | 4.0 |
| pixel_7_6 | 0.0 | 0.0 | 9.0 | 0.0 | 0.0 |
| pixel_7_7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| number_label | 0.0 | 1.0 | 2.0 | 3.0 | 4.0 |
# drop label column
X_digits = digits_df.drop('number_label', axis=1)
digits_labels = digits_df['number_label']
# select a single images
img_idx = 333
Single_Digit = np.array(X_digits.iloc[img_idx])
Single_Digit.shape
# the images inside the dataset are flattened
# (64,)
# need to be turned back into their 8x8 pixel format
Single_Digit = Single_Digit.reshape((8, 8))
Single_Digit.shape
# (8, 8)
# Display the Image
plt.figure(figsize=(4,4))
plt.imshow(Single_Digit, interpolation='nearest', cmap='plasma')
plt.title('Digit Label: %d' % digits_labels[img_idx])
plt.show()
plt.figure(figsize=(8,6))
plt.title('Digit Label: %d' % digits_labels[0])
sns.heatmap(
Single_Digit,
linewidth=0.5,
cmap='plasma_r',
annot=True
)
plt.savefig('assets/Scikit_Learn_122.webp', bbox_inches='tight')
Dataset 2 Preprocessing
# normalize data
scaler = StandardScaler()
digits_scaled = pd.DataFrame(
scaler.fit_transform(X_digits), columns=X_digits.columns
)
digits_scaled.head(5).transpose()
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| pixel_0_0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| pixel_0_1 | -0.335016 | -0.335016 | -0.335016 | -0.335016 | -0.335016 |
| pixel_0_2 | -0.043081 | -1.094937 | -1.094937 | 0.377661 | -1.094937 |
| pixel_0_3 | 0.274072 | 0.038648 | -1.844742 | 0.744919 | -2.551014 |
| pixel_0_4 | -0.664478 | 0.268751 | 0.735366 | 0.268751 | -0.197863 |
| ... | |||||
| pixel_7_3 | 0.208293 | -0.249010 | -2.078218 | 0.208293 | -2.306869 |
| pixel_7_4 | -0.366771 | 0.849632 | -0.164037 | 0.241430 | 0.849632 |
| pixel_7_5 | -1.146647 | 0.548561 | 1.565686 | 0.379040 | -0.468564 |
| pixel_7_6 | -0.505670 | -0.505670 | 1.695137 | -0.505670 | -0.505670 |
| pixel_7_7 | -0.196008 | -0.196008 | -0.196008 | -0.196008 | -0.196008 |
Model Fitting
pca_model2 = PCA(n_components=2)
pca_results2 = pca_model2.fit_transform(digits_scaled)
print(np.sum(pca_model2.explained_variance_ratio_))
# reducing the number of dimensions from 64 -> 2 leads to 22% explained variance
X_digits[['PC1','PC2']] = pca_results2
X_digits[['PC1','PC2']].head(5).transpose()
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| PC1 | 1.914264 | 0.588997 | 1.302144 | -3.020847 | 4.528854 |
| PC2 | -0.954564 | 0.924622 | -0.317291 | -0.868696 | -1.093369 |
plt.figure(figsize=(12,5))
plt.title('PCA Digits Dataset - Coloured by Labels')
sns.scatterplot(
data=X_digits,
x='PC1', y='PC2',
hue=digits_labels,
palette='tab20'
)
plt.legend(bbox_to_anchor=(1.01,1.01))
plt.savefig('assets/Scikit_Learn_123.webp', bbox_inches='tight')
# numbers 4 and 7 are very distinct. There is some overlap between 6 and 0 and between 2 and 3
# but you can still get some separation. All the numbers in the middle are 'problematic' and
# probably need a larger amount training data.
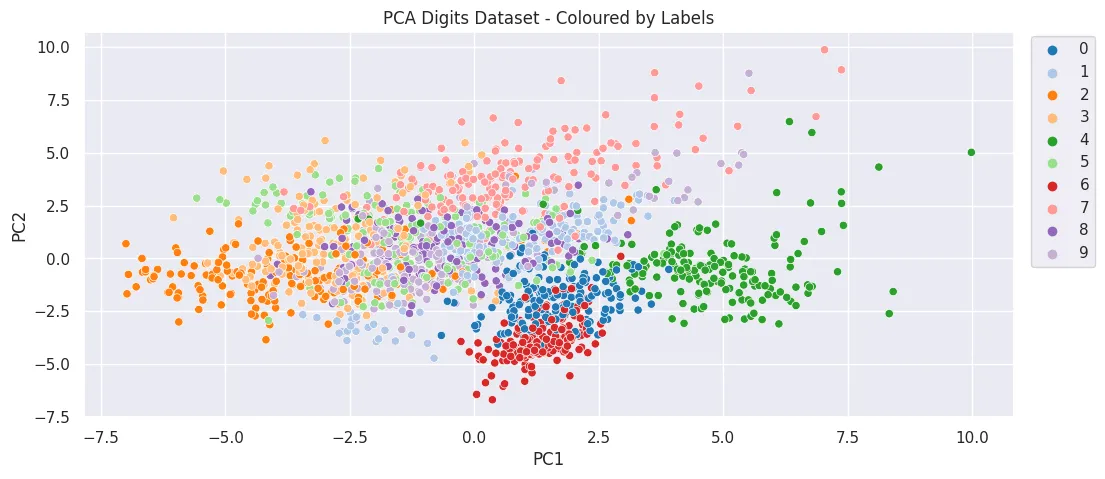
# how many components would we have to add to reach 80% explained variance
explained_variance = []
for n in range(1,65):
pca = PCA(n_components=n)
pca.fit(digits_scaled)
explained_variance.append(np.sum(pca.explained_variance_ratio_))
plt.figure(figsize=(16, 5))
plt.title('Explained Variance by Number of Principal Components')
plt.xlabel('Principal Components')
sns.set(style='darkgrid')
sns.barplot(
data=pd.DataFrame(explained_variance, columns=['Explained Variance']),
x=np.arange(1,65),
y='Explained Variance'
)
plt.savefig('assets/Scikit_Learn_124.webp', bbox_inches='tight')
# we need more than 20 principal components out of 64 to reach 80% expainable variance:
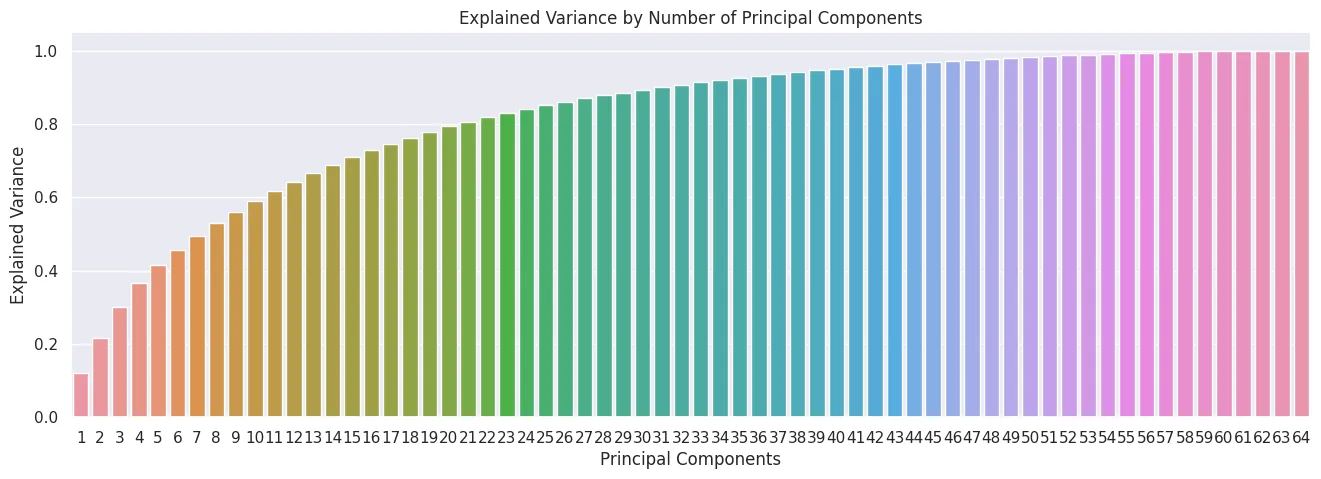
# rerun the training for 3 components for ~30% explained variance
pca_model3 = PCA(n_components=3)
pca_results3 = pca_model3.fit_transform(digits_scaled)
print(np.sum(pca_model3.explained_variance_ratio_))
# reducing the number of dimensions from 64 -> 3 leads to 30% explained variance
X_digits[['PC1','PC2','PC3']] = pca_results3
X_digits[['PC1','PC2','PC3']].head(5).transpose()
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| PC1 | 1.914213 | 0.588981 | 1.302030 | -3.020765 | 4.528946 |
| PC2 | -0.954510 | 0.924646 | -0.317199 | -0.868788 | -1.093498 |
| PC3 | -3.945982 | 3.924713 | 3.023435 | -0.801779 | 0.973213 |
%matplotlib notebook
fig = plt.figure(figsize=(8,8))
ax = plt.axes(projection='3d')
ax.scatter3D(
xs=X_digits['PC1'],
ys=X_digits['PC2'],
zs=X_digits['PC3'],
c=digits_labels,
cmap='tab20'
)
ax.set_title('PCA Digits Dataset - Coloured by Labels')
ax.set(
xticklabels=[],
yticklabels=[],
zticklabels=[],
xlabel='PC1',
ylabel='PC2',
zlabel='PC3',
)
# plt.savefig('assets/Scikit_Learn_125.webp', bbox_inches='tight')