Multidimensional Scaling is a family of statistical methods that focus on creating mappings of items based on distance.
Dimensionality Reduction
Manifold learning is an approach to non-linear dimensionality reduction. Algorithms for this task are based on the idea that the dimensionality of many data sets is only artificially high.
High-dimensional datasets can be very difficult to visualize. While data in two or three dimensions can be plotted to show the inherent structure of the data, equivalent high-dimensional plots are much less intuitive. To aid visualization of the structure of a dataset, the dimension must be reduced in some way.
The simplest way to accomplish this dimensionality reduction is by taking a random projection of the data. Though this allows some degree of visualization of the data structure, the randomness of the choice leaves much to be desired. In a random projection, it is likely that the more interesting structure within the data will be lost.
To address this concern, a number of supervised and unsupervised linear dimensionality reduction frameworks have been designed, such as:
- Principal Component Analysis (PCA)
- Locally Linear Embedding (LLE)
- tStochastic Neighbor Embedding (t-SNE)
- Isometric Mapping (ISOMAP)
- Fisher Linear Discriminant Analysis (LDA)
Dataset
A multivariate study of variation in two species of rock crab of genus Leptograpsus: A multivariate approach has been used to study morphological variation in the blue and orange-form species of rock crab of the genus Leptograpsus. Objective criteria for the identification of the two species are established, based on the following characters:
- SP: Species (Blue or Orange)
- Sex: Male or Female
- FL: Width of the frontal region of the carapace;
- RW: Width of the posterior region of the carapace (rear width);
- CL: Length of the carapace along the midline;
- CW: Maximum width of the carapace;
- BD: and the depth of the body;
The dataset can be downloaded from Github.
(see introduction in: Principal Component Analysis PCA)
raw_data = pd.read_csv('data/A_multivariate_study_of_variation_in_two_species_of_rock_crab_of_genus_Leptograpsus.csv')
data = raw_data.rename(columns={
'sp': 'Species',
'sex': 'Sex',
'index': 'Index',
'FL': 'Frontal Lobe',
'RW': 'Rear Width',
'CL': 'Carapace Midline',
'CW': 'Maximum Width',
'BD': 'Body Depth'})
data['Species'] = data['Species'].map({'B':'Blue', 'O':'Orange'})
data['Sex'] = data['Sex'].map({'M':'Male', 'F':'Female'})
data['Class'] = data.Species + data.Sex
data_columns = ['Frontal Lobe',
'Rear Width',
'Carapace Midline',
'Maximum Width',
'Body Depth']
# generate a class variable for all 4 classes
data['Class'] = data.Species + data.Sex
print(data['Class'].value_counts())
data.head(5)
- BlueMale:
50 - BlueFemale:
50 - OrangeMale:
50 - OrangeFemale:
50
| Species | Sex | Index | Frontal Lobe | Rear Width | Carapace Midline | Maximum Width | Body Depth | Class | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Blue | Male | 1 | 8.1 | 6.7 | 16.1 | 19.0 | 7.0 | BlueMale |
| 1 | Blue | Male | 2 | 8.8 | 7.7 | 18.1 | 20.8 | 7.4 | BlueMale |
| 2 | Blue | Male | 3 | 9.2 | 7.8 | 19.0 | 22.4 | 7.7 | BlueMale |
| 3 | Blue | Male | 4 | 9.6 | 7.9 | 20.1 | 23.1 | 8.2 | BlueMale |
| 4 | Blue | Male | 5 | 9.8 | 8.0 | 20.3 | 23.0 | 8.2 | BlueMale |
# normalize data columns
data_norm = data.copy()
data_norm[data_columns] = MinMaxScaler().fit_transform(data[data_columns])
data_norm.describe()
| Index | Frontal Lobe | Rear Width | Carapace Midline | Maximum Width | Body Depth | |
|---|---|---|---|---|---|---|
| count | 200.000000 | 200.000000 | 200.000000 | 200.000000 | 200.000000 | 200.000000 |
| mean | 25.500000 | 0.527233 | 0.455365 | 0.529043 | 0.515053 | 0.511645 |
| std | 14.467083 | 0.219832 | 0.187835 | 0.216382 | 0.209919 | 0.220953 |
| min | 1.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 13.000000 | 0.358491 | 0.328467 | 0.382219 | 0.384000 | 0.341935 |
| 50% | 25.500000 | 0.525157 | 0.459854 | 0.528875 | 0.525333 | 0.503226 |
| 75% | 38.000000 | 0.682390 | 0.569343 | 0.684650 | 0.664000 | 0.677419 |
| max | 50.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
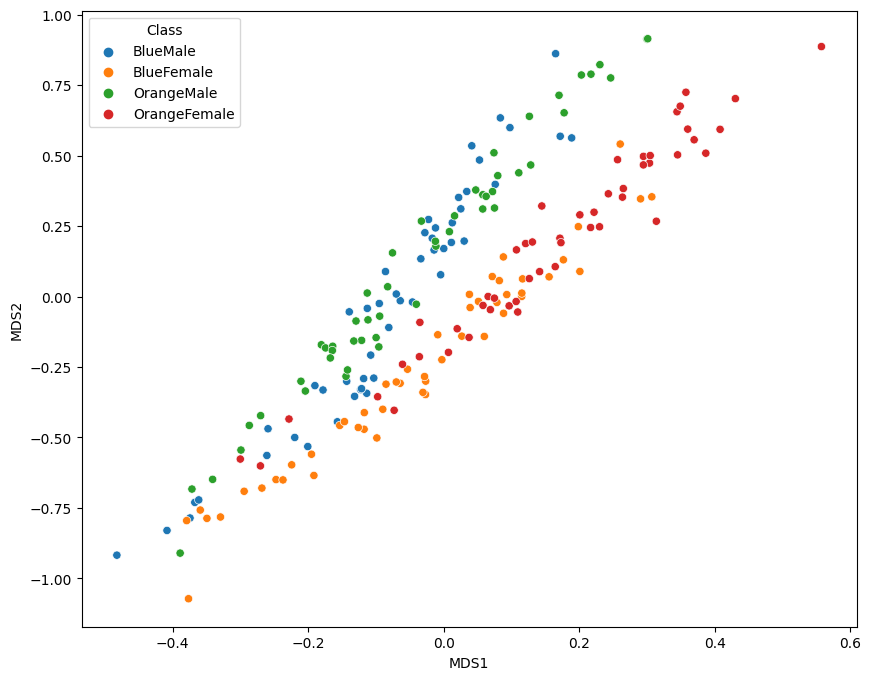
2-Dimensional Plot
no_components = 2
n_init = 15
metric = True
n_stress='auto'
mds = MDS(
n_components=no_components,
n_init=n_init, metric=metric,
normalized_stress=n_stress)
data_mds = mds.fit_transform(data_norm[data_columns])
print('MSE: ', mds.stress_)
# MSE: 3.886582480465905
# the more components you add the smaller
# the mean squared error becomes - meaning
# your model starts to fit better
data_norm[['MDS1', 'MDS2']] = data_mds
data_norm.head(1)
| Species | Sex | Index | Frontal Lobe | Rear Width | Carapace Midline | Maximum Width | Body Depth | Class | MDS1 | MDS2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Blue | Male | 1 | 0.056604 | 0.014599 | 0.042553 | 0.050667 | 0.058065 | BlueMale | -0.482199 | -0.917839 |
fig = plt.figure(figsize=(10, 8))
sns.scatterplot(x='MDS1', y='MDS2', hue='Class', data=data_norm)
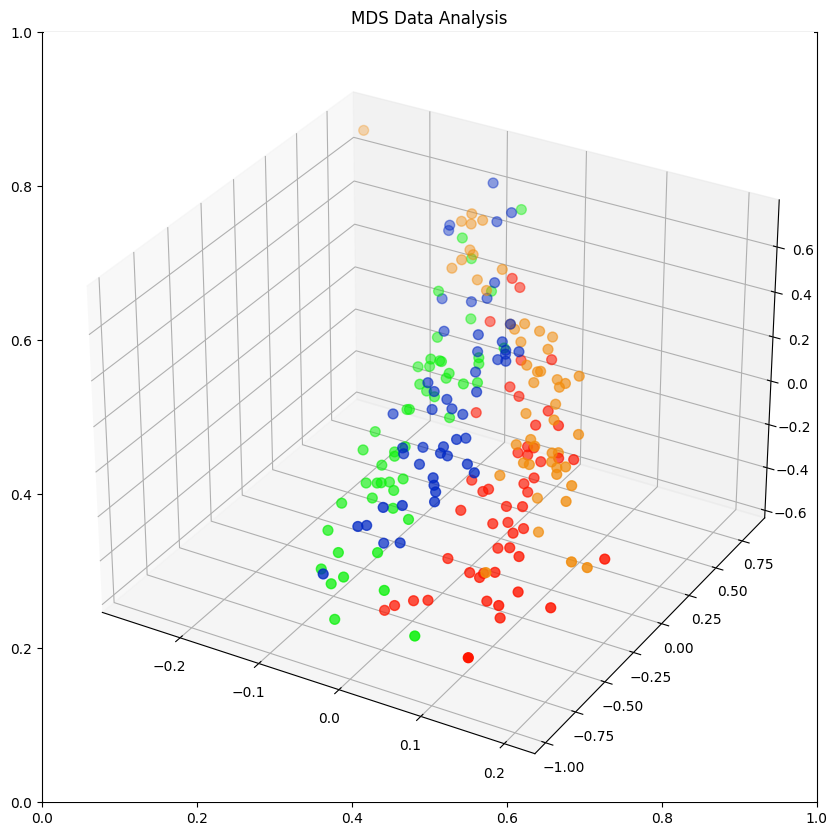
3-Dimensional Plot
no_components = 3
n_init = 15
metric = True
n_stress='auto'
mds = MDS(
n_components=no_components,
n_init=n_init, metric=metric,
normalized_stress=n_stress)
data_mds = mds.fit_transform(data_norm[data_columns])
print('MSE: ', mds.stress_)
# MSE: 2.4601741009431457
data_norm[['MDS1', 'MDS2', 'MDS3']] = data_mds
data_norm.head(1)
| Species | Sex | Index | Frontal Lobe | Rear Width | Carapace Midline | Maximum Width | Body Depth | Class | MDS1 | MDS2 | MDS3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Blue | Male | 1 | 0.056604 | 0.014599 | 0.042553 | 0.050667 | 0.058065 | BlueMale | -0.093961 | 0.80491 | 0.645809 |
class_colours = {
'BlueMale': '#0027c4', #blue
'BlueFemale': '#f18b0a', #orange
'OrangeMale': '#0af10a', # green
'OrangeFemale': '#ff1500', #red
}
colours = data_norm['Class'].apply(lambda x: class_colours[x])
x=data_norm.MDS1
y=data_norm.MDS2
z=data_norm.MDS3
fig = plt.figure(figsize=(10,10))
plt.title('MDS Data Analysis')
ax = fig.add_subplot(projection='3d')
ax.scatter(xs=x, ys=y, zs=z, s=50, c=colours)